1L7X
 
 | | Human liver glycogen phosphorylase b complexed with caffeine, N-acetyl-beta-D-glucopyranosylamine, and CP-403,700 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CAFFEINE, Glycogen phosphorylase, ... | | Authors: | Ekstrom, J.L, Pauly, T.A, Carty, M.D, Soeller, W.C, Culp, J, Danley, D.E, Hoover, D.J, Treadway, J.L, Gibbs, E.M, Fletterick, R.J, Day, Y.S.N, Myszka, D.G, Rath, V.L. | | Deposit date: | 2002-03-18 | | Release date: | 2002-12-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-activity analysis of the purine binding
site of human liver glycogen phosphorylase.
Chem.Biol., 9, 2002
|
|
7RFA
 
 | |
3LK0
 
 | | X-ray structure of bovine SC0067,Ca(2+)-S100B | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CALCIUM ION, Protein S100-B | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int J High Throughput Screen, 2010, 2010
|
|
6UFJ
 
 | | Pistol ribozyme product crystal structure | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*CP*AP*G)-3'), RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(23G))-3'), ... | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6O68
 
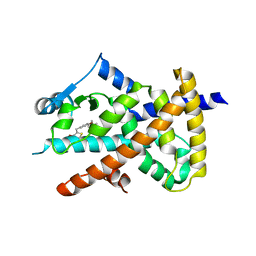 | |
6O7E
 
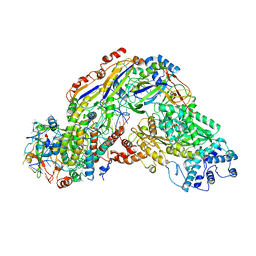 | |
4C2Y
 
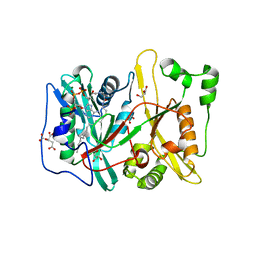 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA co-factor | | Descriptor: | CITRIC ACID, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE 1, ... | | Authors: | Thinon, E, Serwa, R.A, Brannigan, J.A, Brassat, U, Wright, M.H, Heal, W.P, Wilkinson, A.J, Mann, D.J, Tate, E.W. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Global Profiling of Co- and Post-Translationally N-Myristoylated Proteomes in Human Cells.
Nat.Commun., 5, 2014
|
|
4YB1
 
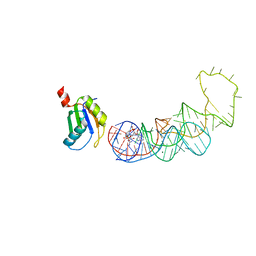 | | 20A Mutant c-di-GMP Vc2 Riboswitch bound with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, RNA (91-MER), ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
3LSR
 
 | |
6UEY
 
 | | Pistol ribozyme transition-state analog vanadate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*UP*CP*UP*GP*CP*UP*CP*UP*CP*(GVA)*UP*CP*CP*AP*A)-3'), RNA (50-MER) | | Authors: | Teplova, M, Falschlunger, C, Krasheninina, O, Patel, D.J, Micura, R. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crucial Roles of Two Hydrated Mg2+Ions in Reaction Catalysis of the Pistol Ribozyme.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4PE1
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC124 | | Descriptor: | CALCIUM ION, DIETHYLCARBAMODITHIOIC ACID, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, A.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
6OJH
 
 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine, ACETATE ION, Biotin carboxylase, ... | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine
To Be Published
|
|
1C49
 
 | | STRUCTURAL AND FUNCTIONAL DIFFERENCES OF TWO TOXINS FROM THE SCORPION PANDINUS IMPERATOR | | Descriptor: | TOXIN K-BETA | | Authors: | Klenk, K.C, Tenenholz, T.C, Matteson, D.R, Rogowski, R.S, Blaustein, M.P, Weber, D.J. | | Deposit date: | 1999-08-17 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural and functional differences of two toxins from the scorpion Pandinus imperator.
Proteins, 38, 2000
|
|
4A7L
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 1) | | Descriptor: | ACTIN, ALPHA SKELETON MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
5ABF
 
 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S,3R,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6OJS
 
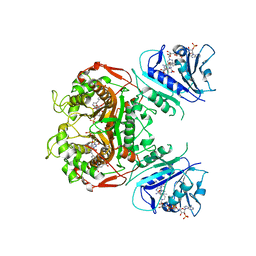 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP, MTX and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-D-glutamic acid | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, ... | | Authors: | Czyzyk, D.J, Anderson, K.S, Jorgensen, W.L, Valhondo, M. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
4ZZA
 
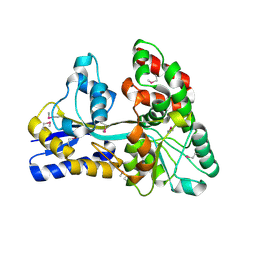 | | Raffinose and panose binding protein from Bifidobacterium animalis subsp. lactis Bl-04, bound with raffinose, selenomethionine derivative | | Descriptor: | Sugar binding protein of ABC transporter system, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose-(1-2)-beta-D-fructofuranose | | Authors: | Fredslund, F, Ejby, M, Andersen, J.M, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2015-05-22 | | Release date: | 2016-06-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An ATP Binding Cassette Transporter Mediates the Uptake of alpha-(1,6)-Linked Dietary Oligosaccharides in Bifidobacterium and Correlates with Competitive Growth on These Substrates.
J. Biol. Chem., 291, 2016
|
|
3LSP
 
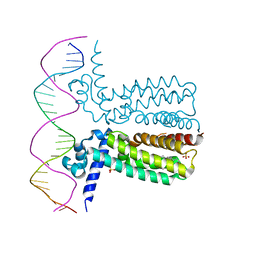 | | Crystal Structure of DesT bound to desCB promoter and oleoyl-CoA | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*TP*CP*GP*AP*GP*TP*CP*AP*AP*CP*AP*AP*GP*CP*GP*TP*TP*CP*AP*CP*TP*GP*AP*TP*GP*TP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*AP*TP*CP*AP*GP*TP*GP*AP*AP*CP*GP*CP*TP*TP*GP*TP*TP*GP*AP*CP*TP*CP*GP*AP*TP*TP*G)-3'), DesT, ... | | Authors: | Miller, D.J, White, S.W. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for the transcriptional regulation of membrane lipid homeostasis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6U7X
 
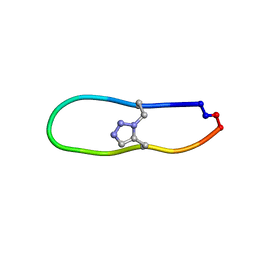 | | NMR solution structure of triazole bridged plasmin inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-TYR-LYS-SER-LYS-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Wang, C.K, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1N1U
 
 | | NMR structure of [Ala1,15]kalata B1 | | Descriptor: | kalata B1 | | Authors: | Daly, N.L, Clark, R.J, Craik, D.J. | | Deposit date: | 2002-10-20 | | Release date: | 2003-02-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Disulfide Folding Pathways of Cystine Knot Proteins. TYING THE KNOT WITHIN THE CIRCULAR BACKBONE OF THE CYCLOTIDES
J.Biol.Chem., 278, 2003
|
|
4PE0
 
 | | Crystal Structure of Calcium-loaded S100B bound to SBi4434 | | Descriptor: | 2-[(2-hydroxyethyl)sulfanyl]naphthalene-1,4-dione, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Pierce, P.D, Wilder, P.T, Neau, D, Toth, E.A, Weber, D.J. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Covalent Small Molecule Inhibitors of Ca(2+)-Bound S100B.
Biochemistry, 53, 2014
|
|
6U9P
 
 | | Wild-type MthK pore in ~150 mM K+ | | Descriptor: | Calcium-gated potassium channel MthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, Nimigean, C.M. | | Deposit date: | 2019-09-09 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Selectivity filter ion binding affinity determines inactivation in a potassium channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1MVJ
 
 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA NMR, 15 STRUCTURES | | Descriptor: | SVIB | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
6O6S
 
 | | Crystal structure of Apo Csm6 | | Descriptor: | Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O7H
 
 | |
