3OD4
 
 | | Crystal Structure of Factor Inhibiting HIF-1 Alpha Complexed with Inhibitor | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | King, O.N.F, Bashford-Rogers, R, Maloney, D.J, Jadhav, A, Heightman, T.D, Simeonov, A, Clifton, I.J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Factor Inhibiting HIF-1 Alpha Complexed with Inhibitor
To be Published
|
|
2F4F
 
 | | Crystal structure of IS200 transposase | | Descriptor: | MANGANESE (II) ION, Transposase, putative | | Authors: | Lee, H.H, Yoon, J.Y, Kim, H.S, Kang, J.Y, Kim, K.H, Kim, D.J, Suh, S.W. | | Deposit date: | 2005-11-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Metal Ion-bound IS200 Transposase
J.Biol.Chem., 281, 2006
|
|
2EUK
 
 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 24:1 Galactosylceramide | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EVT
 
 | | Crystal structure of D48V mutant of human Glycolipid Transfer Protein | | Descriptor: | Glycolipid transfer protein, HEXANE | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
3QZS
 
 | |
2F8T
 
 | |
3RB3
 
 | |
3RBD
 
 | |
3RSD
 
 | |
2HCZ
 
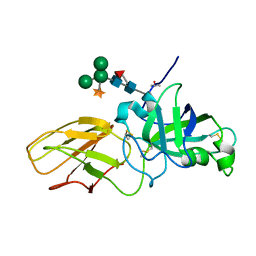 | | Crystal structure of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize | | Descriptor: | Beta-expansin 1a, alpha-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)][alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2006-06-19 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2H0X
 
 | |
3PT6
 
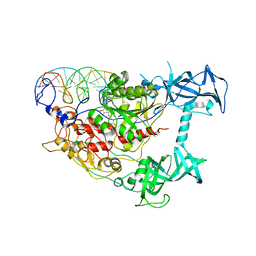 | | Crystal structure of mouse DNMT1(650-1602) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
3QZV
 
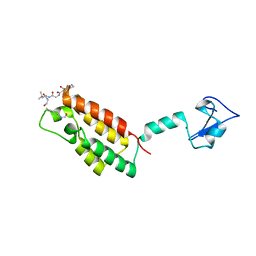 | | Crystal Structure of BPTF PHD-linker-bromo in complex with histone H4K12ac peptide | | Descriptor: | Histone H4, Nucleosome-remodeling factor subunit BPTF, ZINC ION | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
2F8S
 
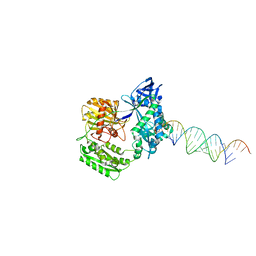 | | Crystal structure of Aa-Ago with externally-bound siRNA | | Descriptor: | 5'-R(P*AP*GP*AP*CP*AP*GP*CP*AP*UP*AP*UP*AP*UP*GP*CP*UP*GP*UP*CP*UP*UP*U)-3', Argonaute protein | | Authors: | Yuan, Y.R, Chen, H.Y, Patel, D.J. | | Deposit date: | 2005-12-03 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Potential Protein-RNA Recognition Event along the RISC-Loading Pathway from the Structure of A. aeolicus Argonaute with Externally Bound siRNA.
Structure, 14, 2006
|
|
2VCA
 
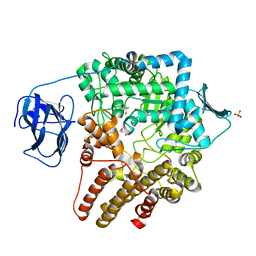 | | Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-N-ACETYLGLUCOSAMINIDASE, CALCIUM ION, ... | | Authors: | Ficko-Blean, E, Stubbs, K.A, Berg, O, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and Mechanistic Insight Into the Basis of Mucopolysaccharidosis Iiib.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2UZR
 
 | | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer (AKT1-PH_E17K) | | Descriptor: | RAC-alpha serine/threonine-protein kinase | | Authors: | Carpten, J.D, Faber, A.L, Horn, C, Donoho, G.P, Briggs, S.L, Robbins, C.M, Hostetter, G, Boguslawski, S, Moses, T.Y, Savage, S, Uhlik, M, Lin, A, Du, J, Qian, Y.W, Zeckner, D.J, Tucker-Kellogg, G, Touchman, J, Patel, K, Mousses, S, Bittner, M, Schevitz, R, Lai, M.H, Blanchard, K.L, Thomas, J.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.
Nature, 448, 2007
|
|
3RIC
 
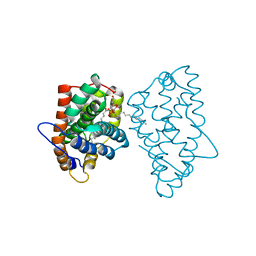 | | Crystal Structure of D48V||A47D mutant of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3PMQ
 
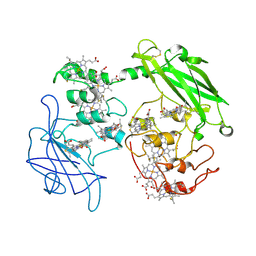 | |
2GMO
 
 | | NMR-structure of an independently folded C-terminal domain of influenza polymerase subunit PB2 | | Descriptor: | Polymerase basic protein 2 | | Authors: | Boudet, J, Tarendeau, F, Guilligay, D, Mas, P, Bougault, C.M, Cusack, S, Simorre, J.-P, Hart, D.J. | | Deposit date: | 2006-04-07 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2H0S
 
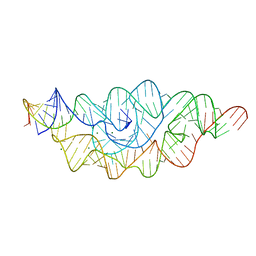 | |
2H4U
 
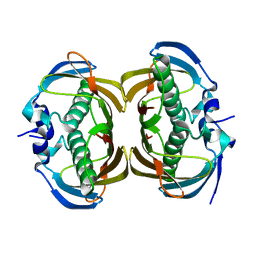 | | Crystal Structure of Human Thioesterase Superfamily Member 2 | | Descriptor: | Thioesterase superfamily member 2 | | Authors: | Ogg, D.J, Uppenberg, J, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Grasslund, S, Flodin, S, Hammerstrom, M, Hogbom, M, Holmberg-Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.-G, Weigelt, J, Hallberg, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of human thioesterase superfamily member 2
To be Published
|
|
3Q0B
 
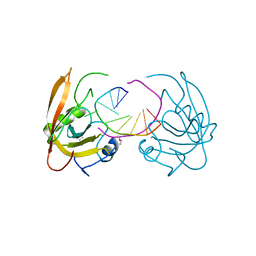 | |
3Q0F
 
 | | Crystal structure of SUVH5 SRA- methylated CHH DNA complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*GP*GP*AP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*(5CM)P*CP*TP*CP*AP*G)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
2GJX
 
 | | Crystallographic structure of human beta-Hexosaminidase A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase alpha chain, ... | | Authors: | Lemieux, M.J, Mark, B.L, Cherney, M.M, Withers, S.G, Mahuran, D.J, James, M.N.G. | | Deposit date: | 2006-03-31 | | Release date: | 2006-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic structure of human beta-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss
of GM2 Ganglioside Hydrolysis
J.Mol.Biol., 359, 2006
|
|
3Q0C
 
 | | Crystal structure of SUVH5 SRA-fully methylated CG DNA complex in space group P6122 | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*(5CM)P*GP*TP*AP*GP*TP*T)-3'), Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH5, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6567 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
