2QHU
 
 | | Structural Basis of Octanoic Acid Recognition by Lipoate-Protein Ligase B | | Descriptor: | Lipoyltransferase, OCTANAL | | Authors: | Kim, D.J, Lee, S.J, Kim, H.S, Kim, K.H, Lee, H.H, Yoon, H.J, Suh, S.W. | | Deposit date: | 2007-07-02 | | Release date: | 2008-02-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of octanoic acid recognition by lipoate-protein ligase B
Proteins, 70, 2008
|
|
2RDZ
 
 | | High Resolution Crystal Structure of the Escherichia coli Cytochrome c Nitrite Reductase. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Hemmings, A.M, RIchardson, D.J. | | Deposit date: | 2007-09-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
2RHU
 
 | |
2RI7
 
 | |
2G8S
 
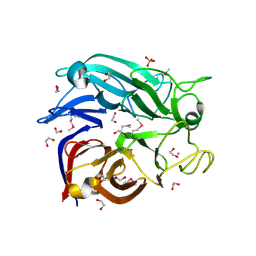 | | Crystal structure of the soluble Aldose sugar dehydrogenase (Asd) from Escherichia coli in the apo-form | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucose/sorbosone dehydrogenases, ... | | Authors: | Southall, S.M, Doel, J.J, Richardson, D.J, Oubrie, A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Soluble Aldose Sugar Dehydrogenase from Escherichia coli: A HIGHLY EXPOSED ACTIVE SITE CONFERRING BROAD SUBSTRATE SPECIFICITY.
J.Biol.Chem., 281, 2006
|
|
2GBW
 
 | | Crystal Structure of Biphenyl 2,3-Dioxygenase from Sphingomonas yanoikuyae B1 | | Descriptor: | Biphenyl 2,3-Dioxygenase Alpha Subunit, Biphenyl 2,3-Dioxygenase Beta Subunit, FE (III) ION, ... | | Authors: | Ferraro, D.J, Brown, E.N, Yu, C, Parales, R.E, Gibson, D.T, Ramaswamy, S. | | Deposit date: | 2006-03-12 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1.
Bmc Struct.Biol., 7, 2007
|
|
2RI5
 
 | |
2RO4
 
 | | RDC-refined Solution Structure of the N-terminal DNA Recognition Domain of the Bacillus subtilis Transition-state Regulator AbrB | | Descriptor: | Transition state regulatory protein abrB | | Authors: | Sullivan, D.M, Bobay, B.G, Kojetin, D.J, Thompson, R.J, Rance, M, Strauch, M.A, Cavanagh, J. | | Deposit date: | 2008-03-08 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insights into the nature of DNA binding of AbrB-like transcription factors
Structure, 16, 2008
|
|
3O7V
 
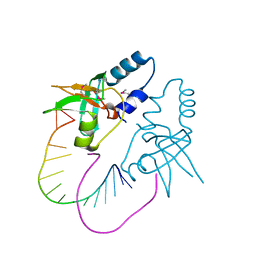 | | Crystal Structure of human Hiwi1 (V361M) PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-08-01 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2RI2
 
 | |
3ODN
 
 | |
2FUI
 
 | |
2FQC
 
 | | Solution structure of conotoxin pl14a | | Descriptor: | Alpha/kappa-conotoxin pl14a | | Authors: | Craik, D.J, Daly, N.L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-07-18 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | A Novel Conotoxin Inhibitor of Kv1.6 Channel and nAChR Subtypes Defines a New Superfamily of Conotoxins
Biochemistry, 45, 2006
|
|
2FUU
 
 | |
3OWW
 
 | |
2RHI
 
 | |
2RHY
 
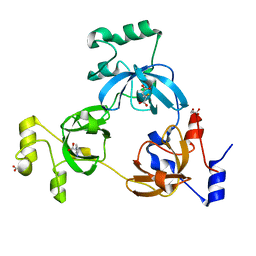 | |
3OXE
 
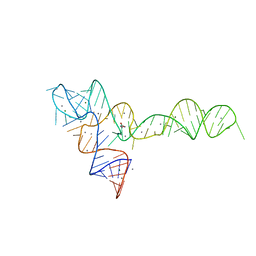 | | crystal structure of glycine riboswitch, Mn2+ soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
2NYA
 
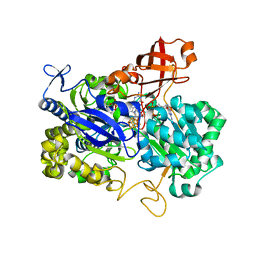 | | Crystal structure of the periplasmic nitrate reductase (NAP) from Escherichia coli | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM(VI) ION, ... | | Authors: | Jepson, B.J.N, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Spectropotentiometric and structural analysis of the periplasmic nitrate reductase from Escherichia coli
J.Biol.Chem., 282, 2007
|
|
2OE5
 
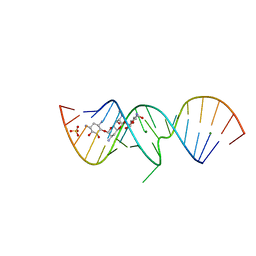 | | 1.5 A X-ray crystal structure of Apramycin complex with RNA fragment GGCGUCGCUAGUACCG/GGUACUAAAAGUCGCCC containing the human ribosomal decoding A site: RNA construct with 3'-overhang | | Descriptor: | APRAMYCIN, MAGNESIUM ION, RNA (5'-R(*GP*GP*CP*GP*UP*CP*GP*CP*UP*AP*GP*UP*AP*CP*CP*G)-3'), ... | | Authors: | Hermann, T, Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2006-12-28 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Apramycin recognition by the human ribosomal decoding site.
Blood Cells Mol.Dis., 38, 2007
|
|
2F2I
 
 | |
2EUM
 
 | | Crystal structure of human Glycolipid Transfer Protein complexed with 8:0 Lactosylceramide | | Descriptor: | DECANE, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
3QRY
 
 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Streptococcus pneumoniae SP_2144 1-deoxymannojirimycin complex | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYMANNOJIRIMYCIN, Putative uncharacterized protein | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-18 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
2F6N
 
 | |
2F33
 
 | | NMR solution structure of Ca2+-loaded calbindin D28K | | Descriptor: | Calbindin | | Authors: | Kojetin, D.J, Venters, R.A, Kordys, D.R, Thompson, R.J, Kumar, R, Cavanagh, J. | | Deposit date: | 2005-11-18 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, binding interface and hydrophobic transitions of Ca(2+)-loaded calbindin-D(28K).
Nat.Struct.Mol.Biol., 13, 2006
|
|
