1DJE
 
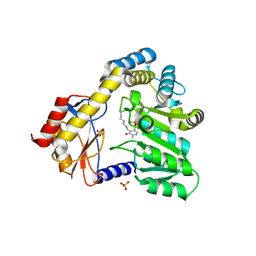 | | CRYSTAL STRUCTURE OF THE PLP-BOUND FORM OF 8-AMINO-7-OXONANOATE SYNTHASE | | Descriptor: | 8-AMINO-7-OXONANOATE SYNTHASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
1IB9
 
 | |
1I34
 
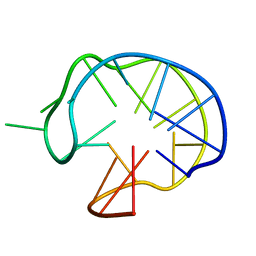 | | SOLUTION DNA QUADRUPLEX WITH DOUBLE CHAIN REVERSAL LOOP AND TWO DIAGONAL LOOPS CONNECTING GGGG TETRADS FLANKED BY G-(T-T) TRIAD AND T-T-T TRIPLE | | Descriptor: | 5'-D(*GP*GP*TP*TP*TP*TP*GP*GP*CP*AP*GP*GP*GP*TP*TP*TP*TP*GP*GP*T)-3' | | Authors: | Kuryavyi, V, Majumdar, A, Shallop, A, Chernichenko, N, Skripkin, E, Jones, R, Patel, D.J. | | Deposit date: | 2001-02-12 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A double chain reversal loop and two diagonal loops define the architecture of a unimolecular DNA quadruplex containing a pair of stacked G(syn)-G(syn)-G(anti)-G(anti) tetrads flanked by a G-(T-T) Triad and a T-T-T triple.
J.Mol.Biol., 310, 2001
|
|
1I39
 
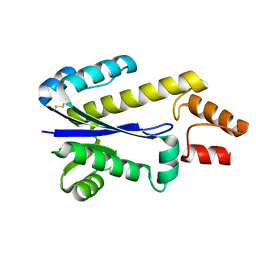 | | RNASE HII FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | RIBONUCLEASE HII | | Authors: | Chapados, B.R, Chai, Q, Hosfield, D.J, Qiu, J, Shen, B, Tainer, J.A. | | Deposit date: | 2001-02-13 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication.
J.Mol.Biol., 307, 2001
|
|
7JW2
 
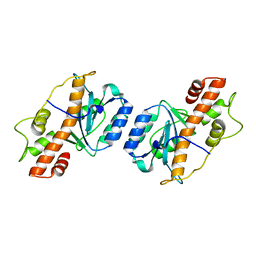 | | Crystal structure of Aedes aegypti Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1DD6
 
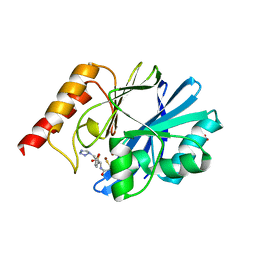 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | Descriptor: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | Authors: | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | Deposit date: | 1999-11-08 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
1DJS
 
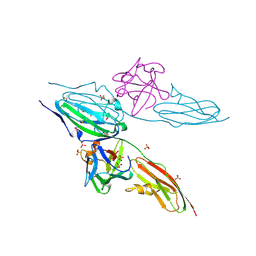 | |
2JSM
 
 | | MONOMERIC HUMAN TELOMERE DNA TETRAPLEX WITH 3+1 STRAND FOLD TOPOLOGY, TWO EDGEWISE LOOPS AND DOUBLE-CHAIN REVERSAL LOOP, NMR, 10 STRUCTURES, Form 1 Natural | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution
Nucleic Acids Res., 35, 2007
|
|
1AT0
 
 | | 17-kDA fragment of hedgehog C-terminal autoprocessing domain | | Descriptor: | 17-HEDGEHOG | | Authors: | Hall, T.M.T, Porter, J.A, Young, K.E, Koonin, E.V, Beachy, P.A, Leahy, D.J. | | Deposit date: | 1997-08-15 | | Release date: | 1997-11-12 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins.
Cell(Cambridge,Mass.), 91, 1997
|
|
1AXU
 
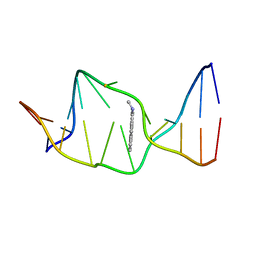 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | Descriptor: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | Authors: | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
1BA4
 
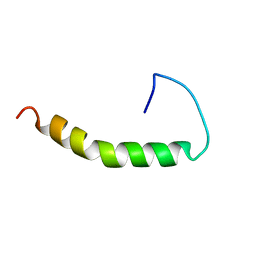 | | THE SOLUTION STRUCTURE OF AMYLOID BETA-PEPTIDE (1-40) IN A WATER-MICELLE ENVIRONMENT. IS THE MEMBRANE-SPANNING DOMAIN WHERE WE THINK IT IS? NMR, 10 STRUCTURES | | Descriptor: | AMYLOID BETA-PEPTIDE | | Authors: | Coles, M, Bicknell, W, Watson, A.A, Fairlie, D.P, Craik, D.J. | | Deposit date: | 1998-04-07 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide(1-40) in a water-micelle environment. Is the membrane-spanning domain where we think it is?
Biochemistry, 37, 1998
|
|
2JMI
 
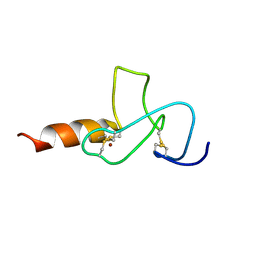 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | Descriptor: | Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
1FSX
 
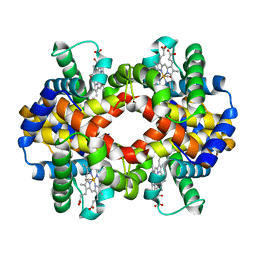 | |
1FA9
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCOGEN PHOSPHORYLASE, LIVER FORM, ... | | Authors: | Rath, V.L, Ammirati, M, LeMotte, P.K, Fennell, K.F, Mansour, M.N, Danley, D.E, Hynes, T.R, Schulte, G.K, Wasilko, D.J, Pandit, J. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core.
Mol.Cell, 6, 2000
|
|
7K7X
 
 | |
1ATJ
 
 | | RECOMBINANT HORSERADISH PEROXIDASE C1A | | Descriptor: | CALCIUM ION, PEROXIDASE C1A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gajhede, M, Schuller, D.J, Henriksen, A, Smith, A.T, Poulos, T.L. | | Deposit date: | 1997-08-14 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of horseradish peroxidase C at 2.15 A resolution.
Nat.Struct.Biol., 4, 1997
|
|
2JCR
 
 | | The hyaluronan binding domain of murine CD44 in a Type B complex with an HA 8-mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
1FJ5
 
 | | TAMOXIFEN-DNA ADDUCT | | Descriptor: | (Z)-2-[4-(1,2)-DIPHENYL-1-BUTENYL)-PHENOXY]-N,N-DIMETHYLETHANAMINIUM, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Shimotakahara, S, Gorin, A, Kolbanovskiy, A, Kettani, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N, Patel, D.J. | | Deposit date: | 2000-08-07 | | Release date: | 2000-09-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Accomodation of S-cis-tamoxifen-N(2)-guanine adduct within a bent and widened DNA minor groove.
J.Mol.Biol., 302, 2000
|
|
2JG2
 
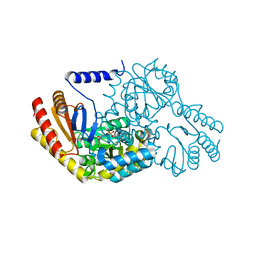 | | HIGH RESOLUTION STRUCTURE OF SPT WITH PLP INTERNAL ALDIMINE | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-05-01 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
2JYY
 
 | |
7K25
 
 | |
7KBH
 
 | |
7JW6
 
 | | Crystal structure of Drosophila Nibbler EXO domain | | Descriptor: | Exonuclease mut-7 homolog | | Authors: | Xie, W, Sowemimo, I, Hayashi, R, Wang, J, Brennecke, J, Ameres, S.L, Patel, D.J. | | Deposit date: | 2020-08-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-function analysis of microRNA 3'-end trimming by Nibbler.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1FT9
 
 | | STRUCTURE OF THE REDUCED (FEII) CO-SENSING PROTEIN FROM R. RUBRUM | | Descriptor: | CARBON MONOXIDE OXIDATION SYSTEM TRANSCRIPTION REGULATOR, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lanzilotta, W.N, Schuller, D.J, Poulos, T.L, Thorsteinsson, M.V, Kirby, B, Roberts, G.P. | | Deposit date: | 2000-09-12 | | Release date: | 2000-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the CO sensing transcription activator CooA.
Nat.Struct.Biol., 7, 2000
|
|
2JSL
 
 | | Monomeric Human Telomere DNA Tetraplex with 3+1 Strand Fold Topology, Two Edgewise Loops and Double-Chain Reversal Loop, Form 2 Natural, NMR, 10 Structures | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-08 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution.
Nucleic Acids Res., 35, 2007
|
|
