1H3B
 
 | | Squalene-Hopene Cyclase | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-{6-[4-(6-BROMO-1,2-BENZISOTHIAZOL-3-YL)PHENOXY]HEXYL}-N-METHYL-2-PROPEN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H9K
 
 | | Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. Phosphate-grown form with tungstate and phosphate bound | | Descriptor: | MOLYBDENUM-BINDING-PROTEIN, PHOSPHATE ION, TUNGSTATE(VI)ION | | Authors: | Delarbre, L, Stevenson, C.E.M, White, D.J, Mitchenall, L.A, Pau, R.N, Lawson, D.M. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein Modg Suggest a Novel Cooperative Binding Mechanism and Provide Insights Into Ligand-Binding Specificity
J.Mol.Biol., 308, 2001
|
|
1H35
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (4'-{[ALLYL(METHYL)AMINO]METHYL}-1,1'-BIPHENYL-4-YL)(4-BROMOPHENYL)METHANONE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H3C
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-(6-{[3-(4-BROMOPHENYL)-1,2-BENZISOTHIAZOL-6-YL]OXY}HEXYL)-N-METHYLPROP-2-EN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1DPS
 
 | | THE CRYSTAL STRUCTURE OF DPS, A FERRITIN HOMOLOG THAT BINDS AND PROTECTS DNA | | Descriptor: | DPS, SODIUM ION | | Authors: | Grant, R.A, Filman, D.J, Finkel, S.E, Kolter, R, Hogle, J.M. | | Deposit date: | 1998-02-23 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Dps, a ferritin homolog that binds and protects DNA.
Nat.Struct.Biol., 5, 1998
|
|
1H39
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ALLYL-{6-[3-(4-BROMO-PHENYL)-1-METHYL-1H-INDAZOL-6-YL]OXY}HEXYL)-N-METHYLAMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H36
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (4-BROMOPHENYL)[4-({(2E)-4-[CYCLOPROPYL(METHYL)AMINO]BUT-2-ENYL}OXY)PHENYL]METHANONE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1H3A
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (2E)-N-ALLYL-4-{[3-(4-BROMOPHENYL)-5-FLUORO-1-METHYL-1H-INDAZOL-6-YL]OXY}-N-METHYL-2-BUTEN-1-AMINE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1GYU
 
 | |
1H37
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE, {4-[((1S,2S)-2-{[ALLYL(CYCLOPROPYL)AMINO]METHYL}CYCLOPROPYL)METHOXY]PHENYL}(4-BROMOPHENYL)METHANONE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1GU6
 
 | | Structure of the Periplasmic Cytochrome c Nitrite Reductase from Escherichia coli | | Descriptor: | CALCIUM ION, CYTOCHROME C552, GLYCEROL, ... | | Authors: | Bamford, V.A, Angove, H.C, Seward, H.E, Thomson, A.J, Cole, J.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Spectroscopy of the Periplasmic Cytochrome C Nitrite Reductase from Escherichia Coli
Biochemistry, 41, 2002
|
|
1GYV
 
 | | Gamma-adaptin appendage domain from clathrin adaptor AP1, L762E mutant | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT | | Authors: | Evans, P.R, Owen, D.J, McMahon, H.M, Kent, H.M. | | Deposit date: | 2002-04-30 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Gamma-adaptin appendage domain: structure and binding site for Eps15 and gamma-synergin.
Structure, 10, 2002
|
|
1H5N
 
 | | DMSO REDUCTASE MODIFIED BY THE PRESENCE OF DMS AND AIR | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM(VI) ION, ... | | Authors: | Bailey, S, Adams, B, Smith, A.T, Richards, R.L, Lowe, D.J, Bray, R.C. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reactions of Dimethyl Sulphoxide Reductase in the Presence of Dimethylsulphide and the Structure of the Dimethylsulphide-Modified Enzyme
Biochemistry, 40, 2001
|
|
1H9O
 
 | | PHOSPHATIDYLINOSITOL 3-KINASE, P85-ALPHA SUBUNIT: C-TERMINAL SH2 DOMAIN COMPLEXED WITH A TYR751 PHOSPHOPEPTIDE FROM THE PDGF RECEPTOR, CRYSTAL STRUCTURE AT 1.79 A | | Descriptor: | BETA-PLATELET-DERIVED GROWTH FACTOR RECEPTOR, PHOSPHATIDYLINOSITOL 3-KINASE | | Authors: | Pauptit, R.A, Rowsell, S, Breeze, A.L, Murshudov, G.N, Dennis, C.A, Derbyshire, D.J, Weston, S.A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | NMR Trial Models: Experiences with the Colicin Immunity Protein Im7 and the P85Alpha C-Terminal Sh2-Peptide Complex
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4TYW
 
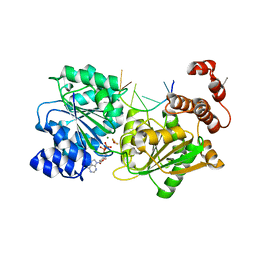 | | DEAD-box helicase Mss116 bound to ssRNA and ADP-BeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase MSS116, mitochondrial, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TZ6
 
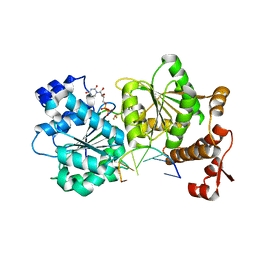 | | DEAD-box helicase Mss116 bound to ssRNA and UDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
1GZI
 
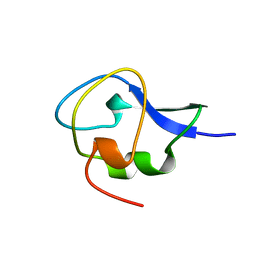 | | CRYSTAL STRUCTURE OF TYPE III ANTIFREEZE PROTEIN FROM OCEAN POUT, AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | HPLC-12 TYPE III ANTIFREEZE PROTEIN | | Authors: | Antson, A.A, Lewis, S, Roper, D.I, Smith, D.J, Hubbard, R.E. | | Deposit date: | 1996-10-25 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Type III Antifreeze Protein from Ocean Pout
To be Published
|
|
4TYY
 
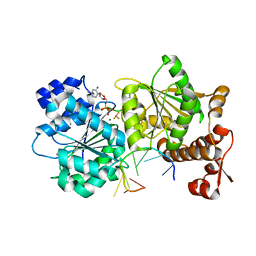 | | DEAD-box helicase Mss116 bound to ssRNA and CDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
1GWC
 
 | | The structure of a tau class glutathione S-transferase from wheat, active in herbicide detoxification | | Descriptor: | GLUTATHIONE S-TRANSFERASE TSI-1, S-HEXYLGLUTATHIONE, SULFATE ION | | Authors: | Thom, R, Cummins, I, Dixon, D.P, Edwards, R, Cole, D.J, Lapthorn, A.J. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a Tau Class Glutathione S-Transferase from Wheat Active in Herbicide Detoxification
Biochemistry, 41, 2002
|
|
1GYW
 
 | | Gamma-adaptin appendage domain from clathrin adaptor AP1 A753D mutant | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT, CHLORIDE ION | | Authors: | Evans, P.R, Miele, A.E, Owen, D.J, McMahon, H.M, Kent, H.M. | | Deposit date: | 2002-04-30 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gamma-adaptin appendage domain: structure and binding site for Eps15 and gamma-synergin.
Structure, 10, 2002
|
|
8G66
 
 | | Structure with SJ3149 | | Descriptor: | (3S)-3-{5-[(1,2-benzoxazol-3-yl)amino]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}piperidine-2,6-dione, Casein kinase I isoform alpha, DNA damage-binding protein 1, ... | | Authors: | Miller, D.J, Young, S.M, Fischer, M. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of ternary complex with molecular glue targeting CK1A for degradation by the CRL4CRBN ubiquitin ligase
To Be Published
|
|
4TYN
 
 | | DEAD-box helicase Mss116 bound to ssDNA and ADP-BeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase MSS116, mitochondrial, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.959 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
1HLK
 
 | | METALLO-BETA-LACTAMASE FROM BACTEROIDES FRAGILIS IN COMPLEX WITH A TRICYCLIC INHIBITOR | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, BETA-LACTAMASE, TYPE II, ... | | Authors: | Payne, D.J, Hueso-Rodriguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Chever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Diez, E, Perez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2000-12-01 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad spectrum inhibitors of metallo-beta-lactamases
Antimicrob.Agents Chemother., 46, 2002
|
|
1HX2
 
 | | SOLUTION STRUCTURE OF BSTI, A TRYPSIN INHIBITOR FROM BOMBINA BOMBINA. | | Descriptor: | BSTI | | Authors: | Rosengren, K.J, Daly, N.L, Scanlon, M.J, Craik, D.J. | | Deposit date: | 2001-01-11 | | Release date: | 2001-01-24 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BSTI: a new trypsin inhibitor from skin secretions of Bombina bombina.
Biochemistry, 40, 2001
|
|
4UN1
 
 | | Sirohaem decarboxylase AhbA/B - an enzyme with structural homology to the Lrp/AsnC transcription factor family that is part of the alternative haem biosynthesis pathway. | | Descriptor: | 12,18-DIDECARBOXY-SIROHEME, PUTATIVE TRANSCRIPTIONAL REGULATOR, ASNC FAMILY | | Authors: | Palmer, D.J, Brown, D.G, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-05-23 | | Release date: | 2014-06-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structure, Function and Properties of Sirohaem Decarboxylase - an Enzyme with Structural Homology to a Transcription Factor Family that is Part of the Alternative Haem Biosynthesis Pathway.
Mol.Microbiol., 93, 2014
|
|
