2ART
 
 | | Crystal structure of lipoate-protein ligase A bound with lipoyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, LIPOIC ACID, Lipoate-protein ligase A, ... | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
2ASJ
 
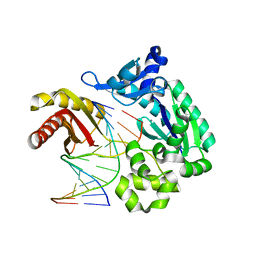 | | oxoG-modified Preinsertion Binary Complex | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*(8OG)*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', CALCIUM ION, ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
1NTB
 
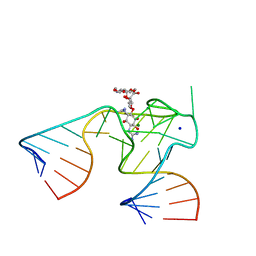 | | 2.9 A crystal structure of Streptomycin RNA-aptamer complex | | Descriptor: | 5'-R(*CP*GP*GP*CP*AP*CP*CP*AP*CP*GP*GP*UP*CP*GP*GP*AP*UP*C)-3', 5'-R(*GP*GP*AP*UP*CP*GP*CP*AP*UP*UP*UP*GP*GP*AP*CP*UP*UP*CP*UP*GP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2003-01-29 | | Release date: | 2003-05-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Encapsulating Streptomycin within a small 40-mer RNA
CHEM.BIOL., 10, 2003
|
|
1NXG
 
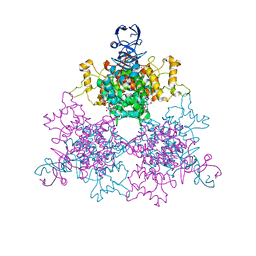 | | The F383A variant of type II Citrate Synthase complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Citrate synthase, SULFATE ION | | Authors: | Maurus, R, Nguyen, N.T, Stokell, D.J, Ayed, A, Hultin, P.G, Duckworth, H.W, Brayer, G.D. | | Deposit date: | 2003-02-10 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the evolution of allosteric properties. The NADH binding site of hexameric type II citrate synthases.
Biochemistry, 42, 2003
|
|
2CWO
 
 | |
1O6H
 
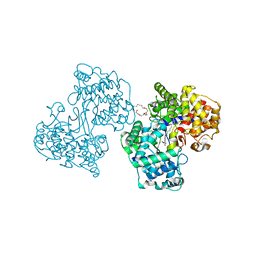 | | Squalene-Hopene Cyclase | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-(6-{[1-(4-BROMOPHENYL)ISOQUINOLIN-6-YL]OXY}HEXYL)-N-METHYLPROP-2-EN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-10-03 | | Release date: | 2003-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
2CHO
 
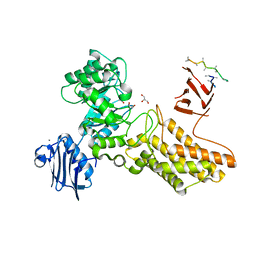 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity | | Descriptor: | ACETATE ION, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
1OWC
 
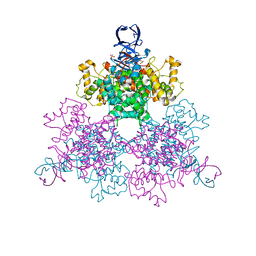 | | Three Dimensional Structure Analysis Of The R109L Variant of the Type II Citrate Synthase From E. Coli | | Descriptor: | Citrate synthase, SULFATE ION | | Authors: | Stokell, D.J, Donald, L.J, Maurus, R, Nguyen, N.T, Sadler, G, Choudhary, K, Hultin, P.G, Brayer, G.D, Duckworth, H.W. | | Deposit date: | 2003-03-28 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the roles of key residues in the unique regulatory NADH binding site of type II citrate synthase of Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
4H9S
 
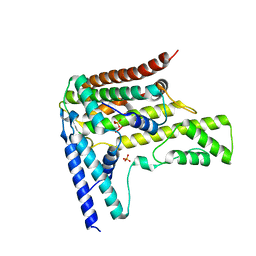 | | Complex structure 6 of DAXX/H3.3(sub7)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
2CO0
 
 | | WDR5 and unmodified Histone H3 complex at 2.25 Angstrom | | Descriptor: | HISTONE H3 DIMETHYL-LYSINE 4, WD-REPEAT PROTEIN 5 | | Authors: | Ruthenburg, A.J, Wang, W, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Histone H3 Recognition and Presentation by the Wdr5 Module of the Mll1 Complex
Nat.Struct.Mol.Biol., 13, 2006
|
|
2ANN
 
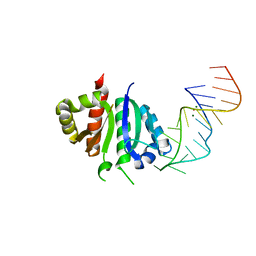 | | Crystal structure (I) of Nova-1 KH1/KH2 domain tandem with 25 nt RNA hairpin | | Descriptor: | 5'-R(*CP*GP*CP*GP*CP*GP*GP*AP*UP*CP*AP*GP*UP*CP*AP*CP*CP*CP*AP*AP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Malinina, L, Teplova, M, Musunuru, K, Teplov, A, Darnell, J.C, Burley, S.K, Darnell, R.B, Patel, D.J. | | Deposit date: | 2005-08-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-RNA and protein-protein recognition by dual KH1/2 domains of the neuronal splicing factor Nova-1.
Structure, 19, 2011
|
|
4GZ6
 
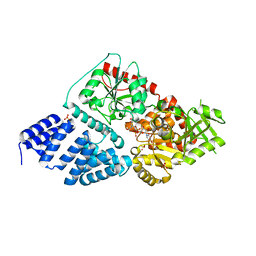 | | Crystal structure of human O-GlcNAc Transferase with UDP-5SGlcNAc | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, SULFATE ION, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Lazarus, M.B, Jiang, J, Gloster, T.M, Zandberg, W.F, Vocadlo, D.J, Walker, S. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural snapshots of the reaction coordinate for O-GlcNAc transferase.
Nat.Chem.Biol., 8, 2012
|
|
2ATL
 
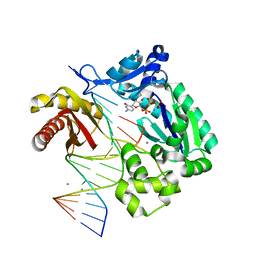 | | Unmodified Insertion Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*T*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-25 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
1OWB
 
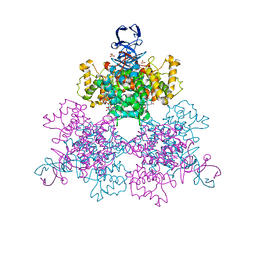 | | Three Dimensional Structure Analysis Of The Variant R109L NADH Complex of Type II Citrate Synthase From E. Coli | | Descriptor: | Citrate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Stokell, D.J, Donald, L.J, Maurus, R, Nguyen, N.T, Sadler, G, Choudhary, K, Hultin, P.G, Brayer, G.D, Duckworth, H.W. | | Deposit date: | 2003-03-28 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the roles of key residues in the unique regulatory NADH binding site of type II citrate synthase of Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
1ONT
 
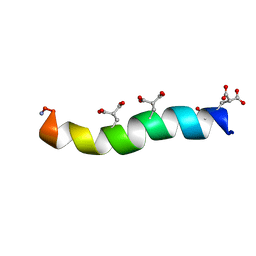 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-T, NMR, 17 STRUCTURES | | Descriptor: | CONANTOKIN-T | | Authors: | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-27 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
1AY1
 
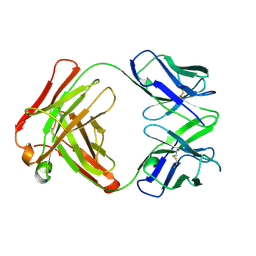 | | ANTI TAQ FAB TP7 | | Descriptor: | TP7 FAB | | Authors: | Murali, R, Helmer-Citterich, M, Sharkey, D.J, Scalice, E.R, Daiss, J.L, Sullivan, M.A, Murthy, H.M.K. | | Deposit date: | 1997-11-13 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural studies on an inhibitory antibody against Thermus aquaticus DNA polymerase suggest mode of inhibition.
Protein Eng., 11, 1998
|
|
1B3P
 
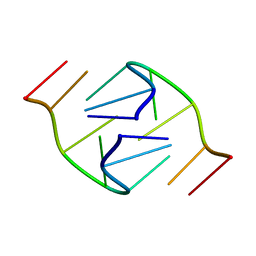 | | 5'-D(*GP*GP*AP*GP*GP*AP*T)-3' | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*AP*T)-3') | | Authors: | Kettani, A, Bouaziz, S, Skripkin, E, Majumdar, A, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-12-14 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Interlocked mismatch-aligned arrowhead DNA motifs.
Structure Fold.Des., 7, 1999
|
|
4H9P
 
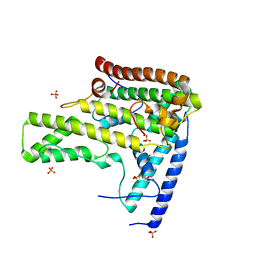 | | Complex structure 3 of DAXX/H3.3(sub5,G90A)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
1B43
 
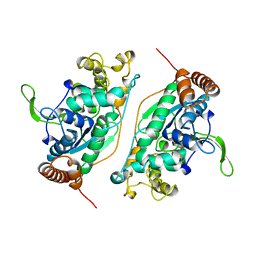 | | FEN-1 FROM P. FURIOSUS | | Descriptor: | PROTEIN (FEN-1) | | Authors: | Hosfield, D.J, Mol, C.D, Shen, B, Tainer, J.A. | | Deposit date: | 1999-01-05 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity.
Cell(Cambridge,Mass.), 95, 1998
|
|
1BA6
 
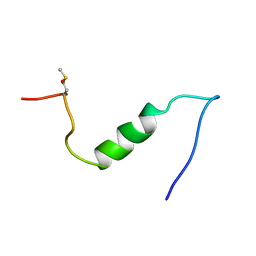 | |
1AT0
 
 | | 17-kDA fragment of hedgehog C-terminal autoprocessing domain | | Descriptor: | 17-HEDGEHOG | | Authors: | Hall, T.M.T, Porter, J.A, Young, K.E, Koonin, E.V, Beachy, P.A, Leahy, D.J. | | Deposit date: | 1997-08-15 | | Release date: | 1997-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins.
Cell(Cambridge,Mass.), 91, 1997
|
|
1OS7
 
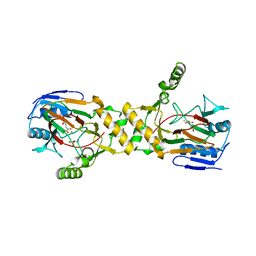 | | Crystal structure of TauD with iron, alpha-ketoglutarate and Taurine bound at pH 7.5 | | Descriptor: | 2-AMINOETHANESULFONIC ACID, 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent taurine dioxygenase, ... | | Authors: | O'Brien, J.R, Schuller, D.J, Yang, V.S, Dillard, B.D, Lanzilotta, W.N. | | Deposit date: | 2003-03-18 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate-Induced Conformational Changes in Escherichia coli Taurine/alpha-Ketoglutarate Dioxygenase and Insight Into the Oligomeric Structure
Biochemistry, 42, 2003
|
|
1OT8
 
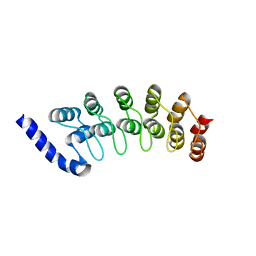 | | Structure of the Ankyrin Domain of the Drosophila Notch Receptor | | Descriptor: | MAGNESIUM ION, Neurogenic locus Notch protein | | Authors: | Zweifel, M.E, Leahy, D.J, Hughson, F.M, Barrick, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and stability of the ankyrin domain of the Drosophila Notch receptor
Protein Sci., 12, 2003
|
|
1OJO
 
 | |
1OM9
 
 | | Structure of the GGA1-appendage in complex with the p56 binding peptide | | Descriptor: | 15-mer peptide fragment of p56, ADP-ribosylation factor binding protein GGA1 | | Authors: | Collins, B.M, Praefcke, G.J.K, Robinson, M.S, Owen, D.J. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of accessory proteins by the appendage domain of GGAs
Nat.Struct.Biol., 10, 2003
|
|
