1R1N
 
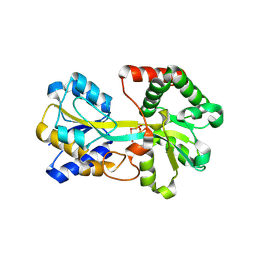 | | Tri-nuclear oxo-iron clusters in the ferric binding protein from N. gonorrhoeae | | Descriptor: | Ferric-iron Binding Protein, OXO-IRON CLUSTER 1, OXO-IRON CLUSTER 2, ... | | Authors: | Zhu, H, Alexeev, D, Hunter, D.J, Campopiano, D.J, Sadler, P.J. | | Deposit date: | 2003-09-24 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Oxo-iron clusters in a bacterial iron-trafficking protein: new roles for a conserved motif.
Biochem.J., 376, 2003
|
|
1H03
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
2X9H
 
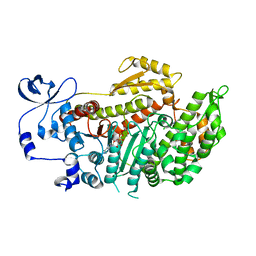 | | CRYSTAL STRUCTURE OF MYOSIN-2 MOTOR DOMAIN IN COMPLEX WITH ADP- METAVANADATE AND PENTACHLOROCARBAZOLE | | Descriptor: | 2,3,4,6,8-PENTACHLORO-9H-CARBAZOL-1-OL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Selvadurai, J, Kirst, J, Knoelker, H.J, Manstein, D.J. | | Deposit date: | 2010-03-19 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Myosin-2 Motor Domain in Complex with Adp-Metavanadate and Pentachlorocarbazole
To be Published
|
|
1W8C
 
 | |
1MWN
 
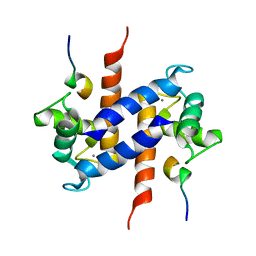 | | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12 | | Descriptor: | CALCIUM ION, F-actin capping protein alpha-1 subunit, S-100 protein, ... | | Authors: | Inman, K.G, Yang, R, Rustandi, R.R, Miller, K.E, Baldisseri, D.M, Weber, D.J. | | Deposit date: | 2002-09-30 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12
J.Mol.Biol., 324, 2002
|
|
8P0L
 
 | | Crystal structure of human O-GlcNAcase in complex with an S-linked CKII peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Protein O-GlcNAcase, ... | | Authors: | Males, A, Davies, G.J, Calvelo, M, Alteen, M.G, Vocadlo, D.J, Rovira, C. | | Deposit date: | 2023-05-10 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human O -GlcNAcase Uses a Preactivated Boat-skew Substrate Conformation for Catalysis. Evidence from X-ray Crystallography and QM/MM Metadynamics.
Acs Catalysis, 13, 2023
|
|
1PIV
 
 | | BINDING OF THE ANTIVIRAL DRUG WIN51711 TO THE SABIN STRAIN OF TYPE 3 POLIOVIRUS: STRUCTURAL COMPARISON WITH DRUG BINDING IN RHINOVIRUS 14 | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, POLIOVIRUS TYPE 3 (SUBUNIT VP1), ... | | Authors: | Hiremath, C.N, Grant, R.A, Filman, D.J, Hogle, J.M. | | Deposit date: | 1995-02-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Binding of the antiviral drug WIN51711 to the sabin strain of type 3 poliovirus: structural comparison with drug binding in rhinovirus 14.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1BU3
 
 | | REFINED CRYSTAL STRUCTURE OF CALCIUM-BOUND SILVER HAKE (PI 4.2) PARVALBUMIN AT 1.65 A. | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN | | Authors: | Richardson, R.C, Nelson, D.J, Royer, W.E, Harrington, D.J. | | Deposit date: | 1998-08-30 | | Release date: | 1999-08-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-Ray crystal structure and molecular dynamics simulations of silver hake parvalbumin (Isoform B).
Protein Sci., 9, 2000
|
|
8J67
 
 | | Crystal structure of Toxoplasma gondii M2AP | | Descriptor: | MIC2-associated protein | | Authors: | Wang, F.F, Zhang, D.J, Zhang, S, Springer, T.A, Song, G.J. | | Deposit date: | 2023-04-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into MIC2 recognition by MIC2-associated protein in Toxoplasma gondii.
Commun Biol, 6, 2023
|
|
8J64
 
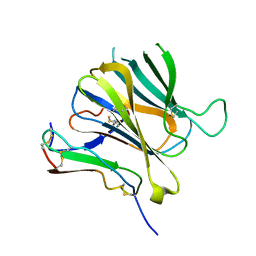 | | Crystal structure of Toxoplasma gondii MIC2-M2AP complex | | Descriptor: | MIC2-associated protein, Micronemal protein MIC2 | | Authors: | Zhang, S, Wang, F.F, Zhang, D.J, Song, G.J, Springer, T.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into MIC2 recognition by MIC2-associated protein in Toxoplasma gondii.
Commun Biol, 6, 2023
|
|
8OQ2
 
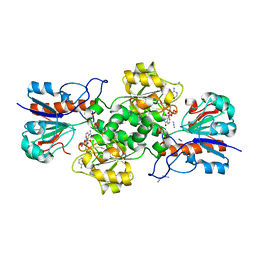 | | Binding of NADP to a formate dehydrogenase from Starkeya novella. | | Descriptor: | AZIDE ION, Formate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Partipilo, M, Whittaker, J.J, Pontillo, N, Guskov, A, Slotboom, D.J. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Binding of NADP to a formate dehydrogenase from Starkeya novella.
To Be Published
|
|
8OVV
 
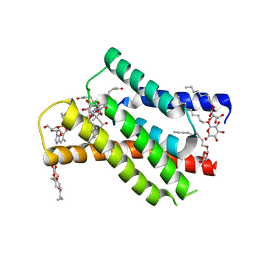 | |
1R9I
 
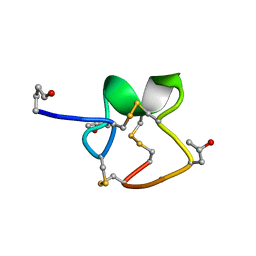 | | NMR Solution Structure of PIIIA toxin, NMR, 20 structures | | Descriptor: | Mu-conotoxin PIIIA | | Authors: | Nielsen, K.J, Watson, M, Adams, D.J, Hammarstrom, A.K, Gage, P.W, Hill, J.M, Craik, D.J, Thomas, L, Adams, D, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2003-10-30 | | Release date: | 2003-11-18 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mu-conotoxin PIIIA, a preferential inhibitor of persistent tetrodotoxin-sensitive sodium channels
J.Biol.Chem., 277, 2002
|
|
1IEO
 
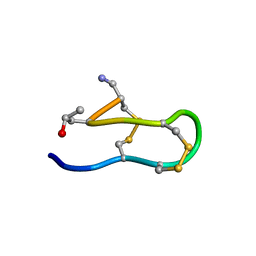 | | SOLUTION STRUCTURE OF MRIB-NH2 | | Descriptor: | PROTEIN MRIB-NH2 | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1OJW
 
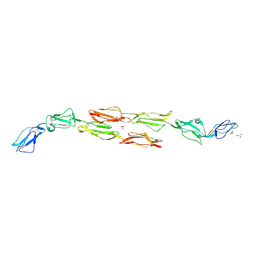 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, SULFATE ION | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, C.M, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8OS2
 
 | | Structure of the human LYVE-1 (lymphatic vessel endothelial receptor-1) hyaluronan binding domain in an unliganded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lymphatic vessel endothelial hyaluronic acid receptor 1 | | Authors: | Ni, T, Bannerji, S, Jackson, D.J, Gilbert, R.J.C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A novel sliding interaction with hyaluronan underlies the mode of action of LYVE-1, the receptor for leucocyte entry and trafficking in the lymphatics
To be published
|
|
1RMK
 
 | | Solution structure of conotoxin MrVIB | | Descriptor: | Mu-O-conotoxin MrVIB | | Authors: | Daly, N.L, Ekberg, J.A, Thomas, L, Adams, D.J, Lewis, R.J, Craik, D.J. | | Deposit date: | 2003-11-28 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structures of muO-conotoxins from Conus marmoreus. Inhibitors of tetrodotoxin (TTX)-sensitive and TTX-resistant sodium channels in mammalian sensory neurons
J.Biol.Chem., 279, 2004
|
|
4DFR
 
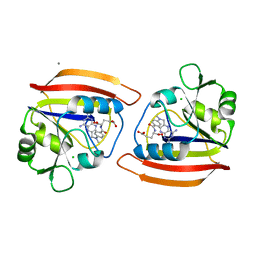 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI AND LACTOBACILLUS CASEI DIHYDROFOLATE REDUCTASE REFINED AT 1.7 ANGSTROMS RESOLUTION. I. GENERAL FEATURES AND BINDING OF METHOTREXATE | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Filman, D.J, Matthews, D.A, Bolin, J.T, Kraut, J. | | Deposit date: | 1982-06-25 | | Release date: | 1982-10-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate.
J.Biol.Chem., 257, 1982
|
|
4DC8
 
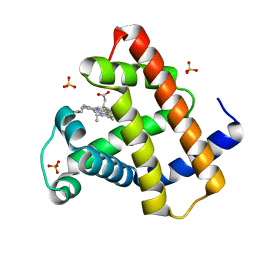 | | Crystal Structure of Myoglobin Unexposed to Excessive SONICC Imaging Laser Dose. | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Becker, M, Mulichak, A.M, Kissick, D.J, Fischetti, R.F, Keefe, L.J, Simpson, D.J. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Towards protein-crystal centering using second-harmonic generation (SHG) microscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1TLL
 
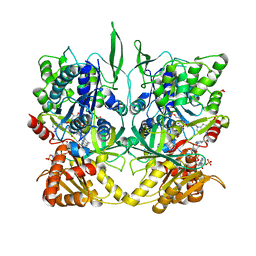 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE REDUCTASE MODULE AT 2.3 A RESOLUTION. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Garcin, E.D, Bruns, C.M, Lloyd, S.J, Hosfield, D.J, Tiso, M, Gachhui, R, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase
J.Biol.Chem., 279, 2004
|
|
2OYT
 
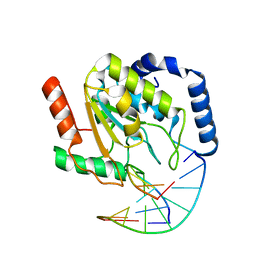 | | Crystal Structure of UNG2/DNA(TM) | | Descriptor: | DNA strand1, DNA strand2, Uracil-DNA glycosylase | | Authors: | Bianchet, M.A, Krosky, D.J, Stivers, J.T, Amzel, L.M. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic capture of an extrahelical thymine in the search for uracil in DNA.
Nature, 449, 2007
|
|
7QZ1
 
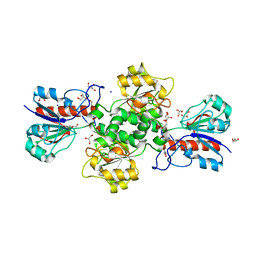 | | Formate dehydrogenase from Starkeya novella | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Pontillo, N, Slotboom, D.J, Guskov, A. | | Deposit date: | 2022-01-30 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural insight into the chemical resistance and cofactor specificity of the formate dehydrogenase from Starkeya novella.
Febs J., 290, 2023
|
|
8PV0
 
 | |
2PF5
 
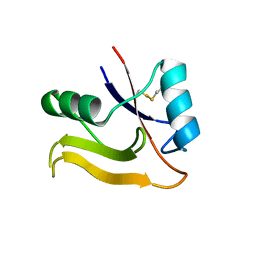 | | Crystal Structure of the Human TSG-6 Link Module | | Descriptor: | NONAETHYLENE GLYCOL, SULFATE ION, Tumor necrosis factor-inducible protein TSG-6 | | Authors: | Higman, V.A, Mahoney, D.J, Noble, M.E.M, Day, A.J. | | Deposit date: | 2007-04-04 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Plasticity of the TSG-6 HA-binding loop and mobility in the TSG-6-HA complex revealed by NMR and X-ray crystallography
J.Mol.Biol., 371, 2007
|
|
8PUZ
 
 | |
