7OG1
 
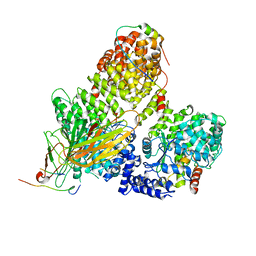 | | AP2 clathrin adaptor core in complex with cargo peptide and FCHO2 | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
7OHO
 
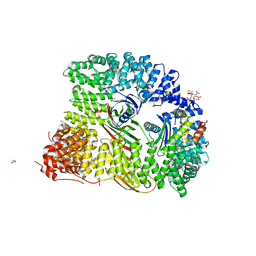 | | Crystal structure of AP2 FCHO2 chimera | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta,F-BAR domain only protein 2, AP-2 complex subunit mu, ... | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
7OIQ
 
 | | Crystal structure of AP2 Mu2 in complex with FCHO2 WxxPhi motif (C2 crystal form) | | Descriptor: | AP-2 complex subunit mu, F-BAR domain only protein 2 | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
7OHZ
 
 | | Crystal structure of AP2 Mu2 - FCHO2 chimera (His6-tagged) | | Descriptor: | AP-2 complex subunit mu,F-BAR domain only protein 2 | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
7OI5
 
 | | Crystal structure of AP2 Mu2 - FCHO2 chimera (GST cleaved) | | Descriptor: | AP-2 complex subunit mu,F-BAR domain only protein 2, GLYCEROL | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
7OFP
 
 | | Apo Structure of Mu2 Adaptin Subunit (Ap50) Of AP2 Clathrin Adaptor | | Descriptor: | AP-2 complex subunit mu, CITRIC ACID, GLYCEROL | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
7OHI
 
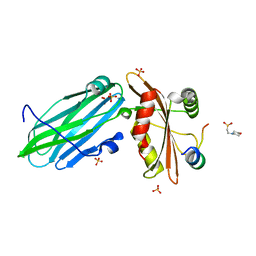 | | FCHO1-peptide-AP2 alpha ear complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AP-2 complex subunit alpha-2, F-BAR domain only protein 1, ... | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
1B9K
 
 | |
1BH4
 
 | |
1HG7
 
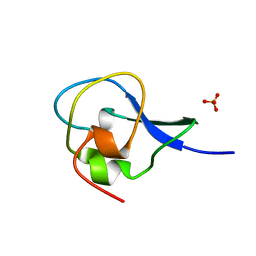 | | High resolution structure of HPLC-12 type III antifreeze protein from Ocean Pout Macrozoarces americanus | | Descriptor: | HPLC-12 TYPE III ANTIFREEZE PROTEIN, SULFATE ION | | Authors: | Antson, A.A, Smith, D.J, Roper, D.I, Lewis, S, Caves, L.S.D, Verma, C.S, Buckley, S.L, Lillford, P.J, Hubbard, R.E. | | Deposit date: | 2000-12-13 | | Release date: | 2001-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Understanding the Mechanism of Ice Binding by Type III Antifreeze Protein
J.Mol.Biol., 305, 2001
|
|
2LMS
 
 | | A single GalNAc residue on Threonine-106 modifies the dynamics and the structure of Interferon alpha-2a around the glycosylation site | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Interferon alpha-2 | | Authors: | Ghasriani, H, Belcourt, P.J.F, Sauve, S, Hodgson, D.J, Gingras, G, Brochu, D, Gilbert, M, Aubin, Y. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A single N-acetylgalactosamine residue at threonine 106 modifies the dynamics and structure of interferon alpha2a around the glycosylation site.
J.Biol.Chem., 288, 2013
|
|
4V8V
 
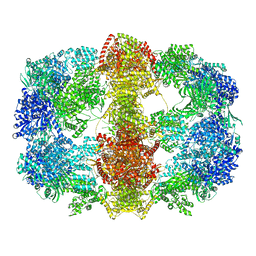 | | Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, TYPE-I FATTY ACID SYNTHASE | | Authors: | Ciccarelli, L, Connell, S.R, Enderle, M, Mills, D.J, Vonck, J, Grininger, M. | | Deposit date: | 2013-04-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure and Conformational Variability of the Mycobacterium Tuberculosis Fatty Acid Synthase Multienzyme Complex.
Structure, 21, 2013
|
|
4W69
 
 | |
4W6D
 
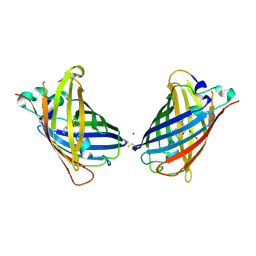 | | Crystal Structure of Full-Length Split GFP Mutant K26C Disulfide Dimer, P 32 2 1 Space Group, Form 1 | | Descriptor: | MAGNESIUM ION, fluorescent protein K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W6I
 
 | |
4W6M
 
 | |
4W6U
 
 | | Crystal Structure of Full-Length Split GFP Mutant E115H/T118H With Nickel Mediated Crystal Contacts, P 21 21 21 Space Group | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, NICKEL (II) ION, ... | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-20 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W77
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 3 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
4W7D
 
 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26H With Copper Mediated Crystal Contacts, P 21 21 21 Space Group | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, COPPER (II) ION, GLYCEROL, ... | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
2LET
 
 | | AN 1H NMR DETERMINATION OF THE THREE DIMENSIONAL STRUCTURES OF MIRROR IMAGE FORMS OF A LEU-5 VARIANT OF THE TRYPSIN INHIBITOR ECBALLIUM ELATERIUM (EETI-II) | | Descriptor: | TRYPSIN INHIBITOR II | | Authors: | Nielsen, K.J, Alewood, D, Andrews, J, Kent, S.B.H, Craik, D.J. | | Deposit date: | 1994-01-04 | | Release date: | 1994-05-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | An 1H NMR determination of the three-dimensional structures of mirror-image forms of a Leu-5 variant of the trypsin inhibitor from Ecballium elaterium (EETI-II).
Protein Sci., 3, 1994
|
|
1H3B
 
 | | Squalene-Hopene Cyclase | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, N-{6-[4-(6-BROMO-1,2-BENZISOTHIAZOL-3-YL)PHENOXY]HEXYL}-N-METHYL-2-PROPEN-1-AMINE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-25 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
1HFO
 
 | |
1AW4
 
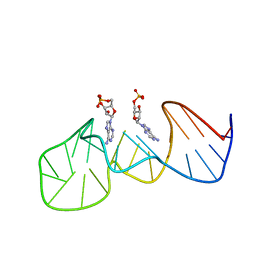 | |
1AX6
 
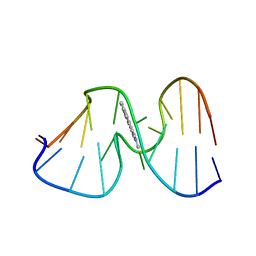 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT OPPOSITE A-2 DELETION SITE IN THE NARI HOT SPOT SEQUENCE CONTEXT; NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(CTCGGC-[AF]G-CCATC)D(GATGGCCGAG) | | Authors: | Mao, B, Gorin, A.A, Gu, Z, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-intercalated conformer of the syn [AF]-C8-dG adduct opposite a--2 deletion site in the NarI hot spot sequence context.
Biochemistry, 36, 1997
|
|
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
