3GIM
 
 | | Dpo4 extension ternary complex with oxoG(anti)-G(syn) pair | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*AP*AP*CP*(8OG)P*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-03-05 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Impact of conformational heterogeneity of OxoG lesions and their pairing partners on bypass fidelity by Y family polymerases.
Structure, 17, 2009
|
|
4A7N
 
 | | Structure of bare F-actin filaments obtained from the same sample as the Actin-Tropomyosin-Myosin Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, F-ACTIN | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
1GWX
 
 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | 2-(4-{3-[1-[2-(2-CHLORO-6-FLUORO-PHENYL)-ETHYL]-3-(2,3-DICHLORO-PHENYL)-UREIDO]-PROPYL}-PHENOXY)-2-METHYL-PROPIONIC ACID, PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-17 | | Release date: | 2000-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
3G5W
 
 | | Crystal structure of Blue Copper Oxidase from Nitrosomonas europaea | | Descriptor: | COPPER (II) ION, CU-O LINKAGE, CU-O-CU LINKAGE, ... | | Authors: | Lawton, T.J, Sayavedra-Soto, L.A, Arp, D.J, Rosenzweig, A.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a two-domain multicopper oxidase: implications for the evolution of multicopper blue proteins.
J.Biol.Chem., 284, 2009
|
|
3LPY
 
 | | Crystal structure of the RRM domain of CyP33 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peptidyl-prolyl cis-trans isomerase E, SULFATE ION | | Authors: | Wang, Z, Patel, D.J. | | Deposit date: | 2010-02-07 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression.
Cell(Cambridge,Mass.), 141, 2010
|
|
4A7H
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 2) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
3G6G
 
 | | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations | | Descriptor: | GLYCEROL, N-{3-[(3-{4-[(4-methoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]-4-methylphenyl}-4-[(4-methylpiperazin-1-yl)methyl]benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Ranjitkar, P, Kasap, C, Shan, Y, Shaw, D.E, Shah, N.P, Kuriyan, J, Maly, D.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations
Cancer Res., 69, 2009
|
|
3LQH
 
 | | Crystal structure of MLL1 PHD3-Bromo in the free form | | Descriptor: | Histone-lysine N-methyltransferase MLL, ZINC ION | | Authors: | Wang, Z, Patel, D.J. | | Deposit date: | 2010-02-09 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression.
Cell(Cambridge,Mass.), 141, 2010
|
|
3LML
 
 | | Crystal Structure of the sheath tail protein Lin1278 from Listeria innocua, Northeast Structural Genomics Consortium Target LkR115 | | Descriptor: | Lin1278 protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target LkR115
To be Published
|
|
3LQJ
 
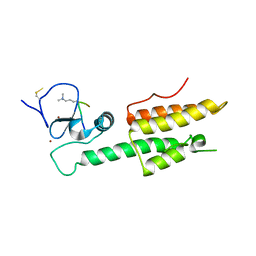 | |
3L6H
 
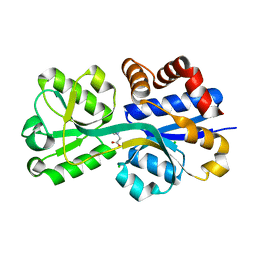 | | Crystal structure of lactococcal OpuAC in its closed-liganded conformation complexed with glycine betaine | | Descriptor: | Betaine ABC transporter permease and substrate binding protein, CHLORIDE ION, TRIMETHYL GLYCINE | | Authors: | Berntsson, R.P.A, Wolters, J.C, Gul, N, Karasawa, A, Thunnissen, A.M.W.H, Slotboom, D.J, Poolman, B. | | Deposit date: | 2009-12-23 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA.
Plos One, 5, 2010
|
|
4CCA
 
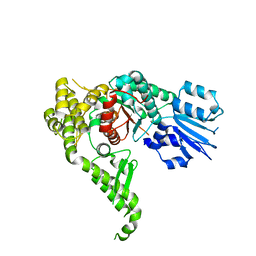 | | Structure of human Munc18-2 | | Descriptor: | CHLORIDE ION, SYNTAXIN-BINDING PROTEIN 2 | | Authors: | Hackmann, Y, Graham, S.C, Ehl, S, Hoening, S, Lehmberg, K, Arico, M, Owen, D.J, Griffiths, G.G. | | Deposit date: | 2013-10-21 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Syntaxin Binding Mechanism and Disease-Causing Mutations in Munc18-2
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4CFV
 
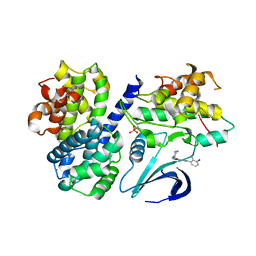 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | Descriptor: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]-2-methylphenol, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | Deposit date: | 2013-11-19 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
3LQI
 
 | |
4BV0
 
 | | High Resolution Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Schuetz, M, Plueckthun, A. | | Deposit date: | 2013-06-24 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3G4D
 
 | | Crystal Structure of (+)-delta-Cadinene Synthase from Gossypium arboreum and Evolutionary Divergence of Metal Binding Motifs for Catalysis | | Descriptor: | (+)-delta-cadinene synthase isozyme XC1, BETA-MERCAPTOETHANOL, GLYCEROL | | Authors: | Gennadios, H.A, Di Costanzo, L, Miller, D.J, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2009-02-03 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis.
Biochemistry, 48, 2009
|
|
3GIO
 
 | | Crystal structure of the TNF-alpha inducing protein (Tip alpha) from Helicobacter pylori | | Descriptor: | Putative uncharacterized protein | | Authors: | Jang, J.Y, Yoon, H.J, Yoon, J.Y, Kim, H.S, Lee, S.J, Kim, K.H, Kim, D.J, Han, B.G, Lee, B.I, Jang, S, Suh, S.W. | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the TNF-alpha-Inducing Protein (Tipalpha) from Helicobacter pylori: Insights into Its DNA-Binding Activity.
J.Mol.Biol., 2009
|
|
3LK1
 
 | | X-ray structure of bovine SC0322,Ca(2+)-S100B | | Descriptor: | 2-sulfanylbenzoic acid, CALCIUM ION, ETHYL MERCURY ION, ... | | Authors: | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | Deposit date: | 2010-01-26 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int J High Throughput Screen, 2010, 2010
|
|
3G6H
 
 | | Src Thr338Ile inhibited in the DFG-Asp-Out conformation | | Descriptor: | N-{4-methyl-3-[(3-{4-[(3,4,5-trimethoxyphenyl)amino]-1,3,5-triazin-2-yl}pyridin-2-yl)amino]phenyl}-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Ranjitkar, P, Kasap, C, Shan, Y, Shaw, D.E, Shah, N.P, Kuriyan, J, Maly, D.J. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations
Cancer Res., 69, 2009
|
|
3LMY
 
 | | The Crystal Structure of beta-hexosaminidase B in complex with Pyrimethamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bateman, K.S, Cherney, M.M, Withers, S.G, Mahuran, D.J, Tropak, M, James, M.N.G. | | Deposit date: | 2010-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of beta-hexosaminidase B in complex with Pyrimethamine
To be Published
|
|
3FU2
 
 | |
4BYF
 
 | | Crystal structure of human Myosin 1c in complex with calmodulin in the pre-power stroke state | | Descriptor: | ADP ORTHOVANADATE, CALMODULIN, MAGNESIUM ION, ... | | Authors: | Munnich, S, Taft, M.H, Pathan-Chhatbar, S, Manstein, D.J. | | Deposit date: | 2013-07-19 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal Structure of Human Myosin 1C-the Motor in Glut4 Exocytosis: Implications for Ca(2+) Regulation and 14-3-3 Binding.
J.Mol.Biol., 426, 2014
|
|
4CC5
 
 | | Fragment-Based Discovery of 6 Azaindazoles As Inhibitors of Bacterial DNA Ligase | | Descriptor: | 2-chloranyl-6-(1H-1,2,4-triazol-3-yl)pyrazine, DNA LIGASE, SULFATE ION | | Authors: | Howard, S, Amin, N, Benowitz, A.B, Chiarparin, E, Cui, H, Deng, X, Heightman, T.D, Holmes, D.J, Hopkins, A, Huang, J, Jin, Q, Kreatsoulas, C, Martin, A.C.L, Massey, F, McCloskey, L, Mortenson, P.N, Pathuri, P, Tisi, D, Williams, P.A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-Based Discovery of 6-Azaindazoles as Inhibitors of Bacterial DNA Ligase.
Acs Med.Chem.Lett., 4, 2013
|
|
4C2Y
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA co-factor | | Descriptor: | CITRIC ACID, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE 1, ... | | Authors: | Thinon, E, Serwa, R.A, Brannigan, J.A, Brassat, U, Wright, M.H, Heal, W.P, Wilkinson, A.J, Mann, D.J, Tate, E.W. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Global Profiling of Co- and Post-Translationally N-Myristoylated Proteomes in Human Cells.
Nat.Commun., 5, 2014
|
|
3LSJ
 
 | |
