4HAN
 
 | | Crystal structure of Galectin 8 with NDP52 peptide | | Descriptor: | Calcium-binding and coiled-coil domain-containing protein 2, DI(HYDROXYETHYL)ETHER, Galectin-8, ... | | Authors: | Kim, B.-W, Hong, S.B, Kim, J.H, Kwon, D.H, Song, H.K. | | Deposit date: | 2012-09-27 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8.
Nat Commun, 4, 2013
|
|
2VBC
 
 | | Crystal structure of the NS3 protease-helicase from Dengue virus | | Descriptor: | DENGUE 4 NS3 FULL-LENGTH PROTEIN, PARTIAL POLYPROTEIN FOR NS2A AND NS2B, TYPE 4 PROTOTYPE DV4 H241 | | Authors: | Luo, D.H, Xu, T, Hunke, C, Gruber, G, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2007-09-10 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of the Ns3 Protease-Helicase from Dengue Virus.
J.Virol., 82, 2008
|
|
4GIQ
 
 | | Crystal Structure of mouse RANK bound to RANKL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, SODIUM ION, ... | | Authors: | Nelson, C.A, Wang, M.W.-H, Fremont, D.H. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RANKL Employs Distinct Binding Modes to Engage RANK and the Osteoprotegerin Decoy Receptor.
Structure, 20, 2012
|
|
7EZM
 
 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gq complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZH
 
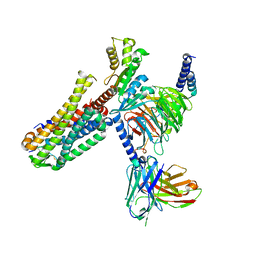 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gi complex | | Descriptor: | Cholecystokinin receptor type A, Cholecystokinin-8, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
7EZK
 
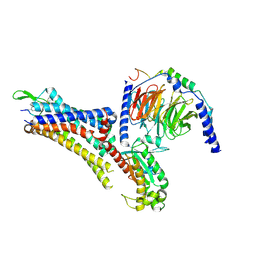 | | Cryo-EM structure of an activated Cholecystokinin A receptor (CCKAR)-Gs complex | | Descriptor: | Chimera of Guanine nucleotide-binding protein G(i) subunit alpha-1 and Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Cholecystokinin receptor type A, Cholecystokinin-8, ... | | Authors: | Liu, Q.F, Yang, D.H, Zhuang, Y.W, Croll, T.I, Cai, X.Q, Duan, J, Dai, A.T, Yin, W.C, Ye, C.Y, Zhou, F.L, Wu, B.L, Zhao, Q, Xu, H.E, Wang, M.W, Jiang, Y. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling selectivity of cholecystokinin A receptor
Nat.Chem.Biol., 17, 2021
|
|
4GQX
 
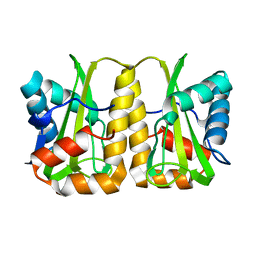 | | Crystal structure of EIIA(NTR) from Burkholderia pseudomallei | | Descriptor: | PTS IIA-like nitrogen-regulatory protein PtsN | | Authors: | Kim, M.-S, Shin, D.H. | | Deposit date: | 2012-08-24 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New molecular interaction of IIA(Ntr) and HPr from Burkholderia pseudomallei identified by X-ray crystallography and docking studies
Proteins, 81, 2013
|
|
4EMG
 
 | | Crystal structure of SpLsm3 | | Descriptor: | Probable U6 snRNA-associated Sm-like protein LSm3 | | Authors: | Jiang, S.M, Wu, D.H, Song, H.W. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe.
Plos One, 7, 2012
|
|
2QRI
 
 | |
7EWJ
 
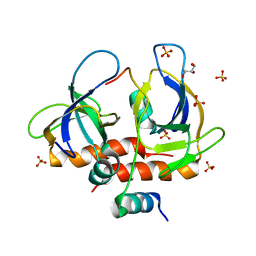 | | Toxin-antitoxin complex from Staphylococcus aureus | | Descriptor: | Endoribonuclease MazF, GLYCEROL, PemI inhibitor, ... | | Authors: | Kim, D.H, Kang, S.M, Lee, S.J, Lee, B.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of PemI in the Staphylococcus aureus PemIK toxin-antitoxin complex: PemI controls PemK by acting as a PemK loop mimic.
Nucleic Acids Res., 50, 2022
|
|
7EWI
 
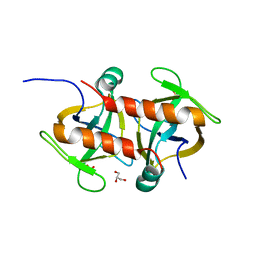 | | Toxin protein from Staphylococcus aureus | | Descriptor: | Endoribonuclease MazF, GLYCEROL, PHOSPHATE ION | | Authors: | Kim, D.H, Kang, S.M, Lee, S.J, Lee, B.J. | | Deposit date: | 2021-05-25 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Role of PemI in the Staphylococcus aureus PemIK toxin-antitoxin complex: PemI controls PemK by acting as a PemK loop mimic.
Nucleic Acids Res., 50, 2022
|
|
7FIN
 
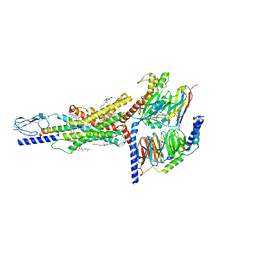 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, GAMMA-L-GLUTAMIC ACID, Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, ... | | Authors: | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-07-31 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7FIY
 
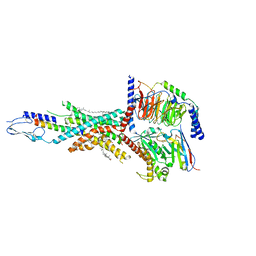 | | Cryo-EM structure of the tirzepatide-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-01 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
7FIM
 
 | | Cryo-EM structure of the tirzepatide (LY3298176)-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,Glucagon-like peptide 1 receptor,human glucagon like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-07-31 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
2VAA
 
 | |
4G59
 
 | |
2QRS
 
 | |
7FEQ
 
 | | Cryo-EM structure of apo BsClpP at pH 6.5 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7FES
 
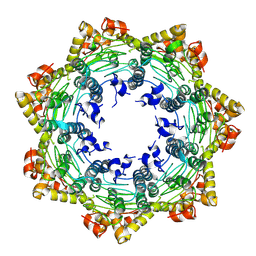 | | Cryo-EM structure of apo BsClpP at pH 4.2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7FER
 
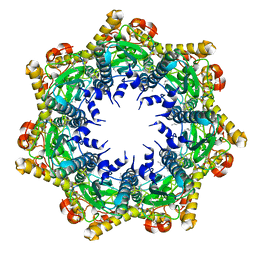 | | Cryo-EM structure of BsClpP-ADEP1 complex at pH 4.2 | | Descriptor: | ADEP1, ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7FEP
 
 | | Cryo-EM structure of BsClpP-ADEP1 complex at pH 6.5 | | Descriptor: | ADEP1, ATP-dependent Clp protease proteolytic subunit | | Authors: | Kim, L, Lee, B.-G, Kim, M.K, Kwon, D.H, Kim, H, Brotz-Oesterhelt, H, Roh, S.-H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-07-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
4EMH
 
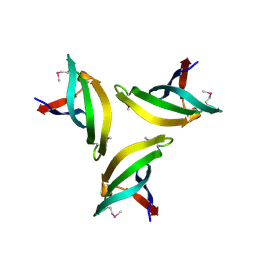 | | Crystal structure of SpLsm4 | | Descriptor: | Probable U6 snRNA-associated Sm-like protein LSm4 | | Authors: | Jiang, S.M, Wu, D.H, Song, H.W. | | Deposit date: | 2012-04-12 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Lsm3, Lsm4 and Lsm5/6/7 from Schizosaccharomyces pombe.
Plos One, 7, 2012
|
|
4EJ0
 
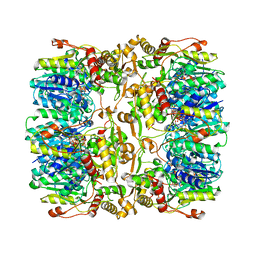 | |
2VVE
 
 | | Crystal structure of the stem and receptor binding domain of the spike protein P1 from bacteriophage PM2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, SPIKE PROTEIN P1 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
2VVF
 
 | | Crystal structure of the major capsid protein P2 from Bacteriophage PM2 | | Descriptor: | CALCIUM ION, MAJOR CAPSID PROTEIN P2 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
