8H0H
 
 | |
4GA1
 
 | |
2QZV
 
 | | Draft Crystal Structure of the Vault Shell at 9 Angstroms Resolution | | Descriptor: | Major vault protein | | Authors: | Anderson, D.H, Kickhoefer, V.A, Sievers, S.A, Rome, L.H, Eisenberg, D. | | Deposit date: | 2007-08-17 | | Release date: | 2007-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (9 Å) | | Cite: | Draft crystal structure of the vault shell at 9-A resolution.
Plos Biol., 5, 2007
|
|
2QRT
 
 | |
4G91
 
 | | CCAAT-binding complex from Aspergillus nidulans | | Descriptor: | HAPB protein, HapE, Transcription factor HapC (Eurofung) | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
4GF2
 
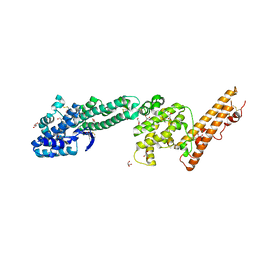 | | Crystal structure of Plasmodium falciparum Erythrocyte Binding Antigen 140 (PfEBA-140/BAEBL) | | Descriptor: | Erythrocyte binding antigen 140, GLYCEROL | | Authors: | Lin, D.H, Malpede, B.M, Batchelor, J.D, Tolia, N.H. | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal and Solution Structures of Plasmodium falciparum Erythrocyte-binding Antigen 140 Reveal Determinants of Receptor Specificity during Erythrocyte Invasion.
J.Biol.Chem., 287, 2012
|
|
8HHD
 
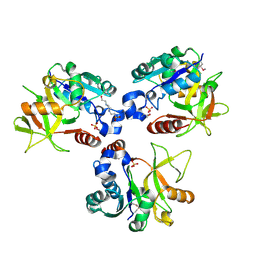 | | Crystal structure of PaMurU | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleotidyl transferase, SULFATE ION | | Authors: | Shin, D.H, Jo, S.R, Kim, M.S. | | Deposit date: | 2022-11-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of PAMurU
To Be Published
|
|
8HIP
 
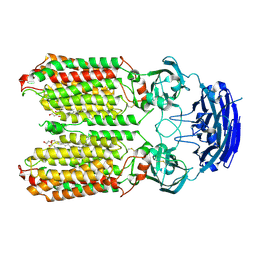 | | dsRNA transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Systemic RNA interference defective protein 1, ZINC ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HKE
 
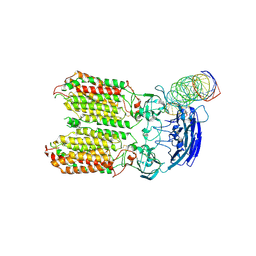 | | dsRNA transporter | | Descriptor: | RNA (37-MER), Systemic RNA interference defective protein 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I1B
 
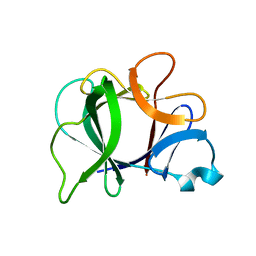 | |
2TSS
 
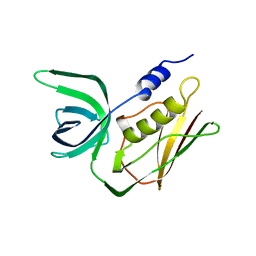 | | TOXIC SHOCK SYNDROME TOXIN-1 FROM STAPHYLOCOCCUS AUREUS: ORTHORHOMBICC222(1) CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-04 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
4DAJ
 
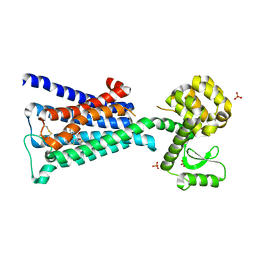 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
4FXB
 
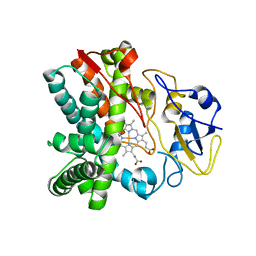 | | Crystal structure of CYP105N1 from Streptomyces coelicolor: a cytochrome P450 oxidase in the coelibactin siderophore biosynthetic pathway | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative cytochrome P450 | | Authors: | Hong, M.K, Lim, Y.R, Kim, J.K, Kim, D.H, Kang, L.W. | | Deposit date: | 2012-07-03 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of cytochrome P450 CYP105N1 from Streptomyces coelicolor, an oxidase in the coelibactin siderophore biosynthetic pathway
Arch.Biochem.Biophys., 528, 2012
|
|
4DD8
 
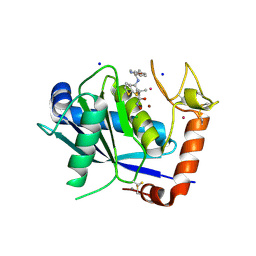 | | ADAM-8 metalloproteinase domain with bound batimastat | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hall, T, Shieh, H.S, Day, J.E, Caspers, N, Chrencik, J.E, Williams, J.M, Pegg, L.E, Pauley, A.M, Moon, A.F, Krahn, J.M, Fischer, D.H, Kiefer, J.R, Tomasselli, A.G, Zack, M.D. | | Deposit date: | 2012-01-18 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ADAM-8 catalytic domain complexed with batimastat.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
7DOP
 
 | | Structural insights into viral RNA capping and plasma membrane targeting by Chikungunya virus nonstructural protein 1 | | Descriptor: | Nonstructural Protein 1, ZINC ION | | Authors: | Zhang, K, Law, Y.S, Law, M.C.Y, Tan, Y.B, Wirawan, M, Luo, D.H. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structural insights into viral RNA capping and plasma membrane targeting by Chikungunya virus nonstructural protein 1.
Cell Host Microbe, 29, 2021
|
|
4GA0
 
 | |
7D34
 
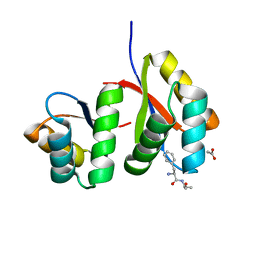 | | AtClpS1-peptide complex | | Descriptor: | ACETIC ACID, ALANINE, ATP-dependent Clp protease adapter protein CLPS1, ... | | Authors: | Heo, J, Kim, L, Kwon, D.H, Song, H.K. | | Deposit date: | 2020-09-18 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for the N-degron specificity of ClpS1 from Arabidopsis thaliana.
Protein Sci., 30, 2021
|
|
4G92
 
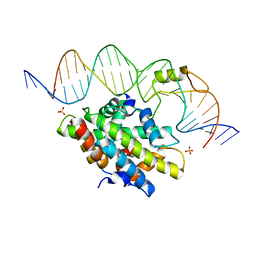 | | CCAAT-binding complex from Aspergillus nidulans with DNA | | Descriptor: | DNA, HAPB protein, HapE, ... | | Authors: | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
2R69
 
 | | Crystal structure of Fab 1A1D-2 complexed with E-DIII of Dengue virus at 3.8 angstrom resolution | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-05 | | Release date: | 2007-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2R5V
 
 | | Hydroxymandelate Synthase Crystal Structure | | Descriptor: | (2S)-hydroxy(4-hydroxyphenyl)ethanoic acid, COBALT (II) ION, PCZA361.1, ... | | Authors: | Brownlee, J.M, He, P, Moran, G.R, Harrison, D.H.T. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two roads diverged: the structure of hydroxymandelate synthase from Amycolatopsis orientalis in complex with 4-hydroxymandelate.
Biochemistry, 47, 2008
|
|
4GA2
 
 | |
4FFY
 
 | | Crystal structure of DENV1-E111 single chain variable fragment bound to DENV-1 DIII, strain 16007. | | Descriptor: | CHLORIDE ION, DENV1-E111 single chain variable fragment (heavy chain), DENV1-E111 single chain variable fragment (light chain), ... | | Authors: | Austin, S.K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Differential Neutralization of DENV-1 Genotypes by an Antibody that Recognizes a Cryptic Epitope.
Plos Pathog., 8, 2012
|
|
7DNJ
 
 | | K63-polyUb MDA5CARDs complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
7DNI
 
 | | MDA5 CARDs-MAVS CARD polyUb complex | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, Mitochondrial antiviral-signaling protein, Ubiquitin | | Authors: | Song, B, Chen, Y, Luo, D.H, Zheng, J. | | Deposit date: | 2020-12-09 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Immunity, 54, 2021
|
|
4FFE
 
 | | The structure of cowpox virus CPXV018 (OMCP) | | Descriptor: | CPXV018 protein | | Authors: | Lazear, E, Peterson, L.W, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Cowpox Virus-Encoded NKG2D Ligand OMCP.
J.Virol., 87, 2013
|
|
