2AQT
 
 | | CU/ZN superoxide dismutase from neisseria meningitidis K91Q, K94Q double mutant | | Descriptor: | COPPER (I) ION, COPPER (II) ION, SULFATE ION, ... | | Authors: | DiDonato, M, Kassmann, C.J, Bruns, C.K, Cabelli, D.E, Cao, Z, Tabatabai, L.B, Kroll, J.S, Getzoff, E.D. | | Deposit date: | 2005-08-18 | | Release date: | 2006-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CU/ZN superoxide dismutase from neisseria meningitidis K91Q, K94Q double mutant
To be Published
|
|
1CMP
 
 | | SMALL MOLECULE BINDING TO AN ARTIFICIALLY CREATED CAVITY AT THE ACTIVE SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Mcree, D.E, Churchill, M.J, Goodin, D.B. | | Deposit date: | 1993-11-23 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small molecule binding to an artificially created cavity at the active site of cytochrome c peroxidase.
Biochemistry, 33, 1994
|
|
2AX0
 
 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5x) | | Descriptor: | 5R-(2E-METHYL-3-PHENYL-ALLYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
2AKW
 
 | | Crystal Structure of T.Thermophilus Phenylalanyl-tRNA synthetase complexed with p-Cl-Phenylalanine | | Descriptor: | 4-CHLORO-L-PHENYLALANINE, MANGANESE (II) ION, Phenylalanyl-tRNA synthetase alpha chain, ... | | Authors: | Kotik-Kogan, O.M, Moor, N.A, Tworowski, D.E, Safro, M.G. | | Deposit date: | 2005-08-04 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Discrimination of L-Phenylalanine from L-Tyrosine by Phenylalanyl-tRNA Synthetase
Structure, 13, 2005
|
|
1PB3
 
 | |
1PB1
 
 | |
2AHQ
 
 | |
2AWZ
 
 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5h) | | Descriptor: | 5R-(4-BROMOPHENYLMETHYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
2ALY
 
 | | Crystal Structure of T.Thermophilus Phenylalanyl-tRNA synthetase complexed with 5'-O-[N-(L-tyrosyl)sulphamoyl]adenosine | | Descriptor: | 5'-O-[N-(L-TYROSYL)SULFAMOYL]ADENOSINE, MANGANESE (II) ION, Phenylalanyl-tRNA synthetase alpha chain, ... | | Authors: | Kotik-Kogan, O.M, Moor, N.A, Tworowski, D.E, Safro, M.G. | | Deposit date: | 2005-08-04 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Discrimination of L-Phenylalanine from L-Tyrosine by Phenylalanyl-tRNA Synthetase
Structure, 13, 2005
|
|
2ERC
 
 | | CRYSTAL STRUCTURE OF ERMC' A RRNA-METHYL TRANSFERASE | | Descriptor: | RRNA METHYL TRANSFERASE | | Authors: | Bussiere, D.E, Muchmore, S.W, Dealwis, C.G, Schluckebier, G, Abad-Zapatero, C. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Crystal structure of ErmC', an rRNA methyltransferase which mediates antibiotic resistance in bacteria.
Biochemistry, 37, 1998
|
|
2F17
 
 | | Mouse Thiamin Pyrophosphokinase in a Ternary Complex with Pyrithiamin Pyrophosphate and AMP at 2.5 angstrom | | Descriptor: | 1-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-2-METHYLPYRIDINIUM, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Liu, J.Y, Timm, D.E, Hurley, T.D. | | Deposit date: | 2005-11-14 | | Release date: | 2005-11-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrithiamine as a substrate for thiamine pyrophosphokinase
J.Biol.Chem., 281, 2006
|
|
1MT1
 
 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannaschii | | Descriptor: | AGMATINE, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE ALPHA CHAIN, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE BETA CHAIN | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1DP0
 
 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | Descriptor: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1999-12-22 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1DC8
 
 | | STRUCTURE OF A TRANSIENTLY PHOSPHORYLATED "SWITCH" IN BACTERIAL SIGNAL TRANSDUCTION | | Descriptor: | NITROGEN REGULATION PROTEIN | | Authors: | Kern, D, Volkman, B.F, Luginbuhl, P, Nohaile, M.J, Kustu, S, Wemmer, D.E. | | Deposit date: | 1999-11-04 | | Release date: | 2000-01-05 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Structure of a transiently phosphorylated switch in bacterial signal transduction.
Nature, 402, 1999
|
|
2E35
 
 | | the minimized average structure of L11 with rg refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
1HC8
 
 | | CRYSTAL STRUCTURE OF A CONSERVED RIBOSOMAL PROTEIN-RNA COMPLEX | | Descriptor: | 58 NUCLEOTIDE RIBOSOMAL 23S RNA DOMAIN, MAGNESIUM ION, OSMIUM ION, ... | | Authors: | Conn, G.L, Draper, D.E, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2001-04-27 | | Release date: | 2002-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact RNA Tertiary Structure Contains a Buried Backbone-K+ Complex
J.Mol.Biol., 318, 2002
|
|
1DC7
 
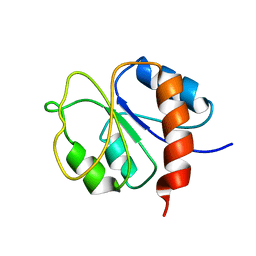 | | STRUCTURE OF A TRANSIENTLY PHOSPHORYLATED "SWITCH" IN BACTERIAL SIGNAL TRANSDUCTION | | Descriptor: | NITROGEN REGULATION PROTEIN | | Authors: | Kern, D, Volkman, B.F, Luginbuhl, P, Nohaile, M.J, Kustu, S, Wemmer, D.E. | | Deposit date: | 1999-11-04 | | Release date: | 2000-01-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a transiently phosphorylated switch in bacterial signal transduction.
Nature, 402, 1999
|
|
1J56
 
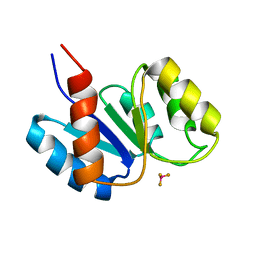 | | MINIMIZED AVERAGE STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURE INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1IBM
 
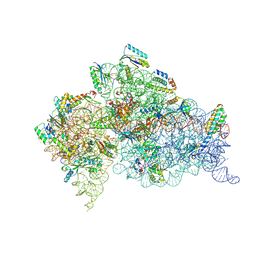 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
1IBL
 
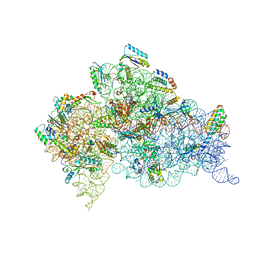 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE AND WITH THE ANTIBIOTIC PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
1IBK
 
 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTIC PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-04 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
1KRX
 
 | | SOLUTION STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURES INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1KRW
 
 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | Descriptor: | NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1IKA
 
 | |
1SI0
 
 | | Crystal Structure of Mannheimia haemolytica Ferric iron-Binding Protein A in a closed conformation | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, FE (III) ION, ... | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, Snell, G, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for iron binding and release by a novel class of periplasmic iron-binding proteins found in gram-negative pathogens.
J.Bacteriol., 186, 2004
|
|
