6REN
 
 | | Crystal structure of 3fPizza6-SH with Zn2+ | | Descriptor: | 3fPizza6-SH, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XX1
 
 | | Low resolution structure of LCAT in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 heavy chain, Fab1 light chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
5ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE, ISOCITRIC ACID, MAGNESIUM ION | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
4XWG
 
 | | Crystal Structure of LCAT (C31Y) in complex with Fab1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1 Heavy Chain, Fab1 Light Chain, ... | | Authors: | Piper, D.E, Walker, N.P.C, Romanow, W.G, Thibault, S.T. | | Deposit date: | 2015-01-28 | | Release date: | 2015-07-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The high-resolution crystal structure of human LCAT.
J.Lipid Res., 56, 2015
|
|
3NV0
 
 | | Crystal structure and mutational analysis of the NXF2/NXT1 heterodimeric complex from caenorhabditis elegans at 1.84 A resolution | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kerkow, D.E, Carmel, A.B, Williamson, J.R. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The structure of the NXF2/NXT1 heterodimeric complex reveals the combined specificity and versatility of the NTF2-like fold.
J.Mol.Biol., 415, 2012
|
|
5IH9
 
 | | MELK in complex with NVS-MELK8A | | Descriptor: | 1-methyl-4-[4-(4-{3-[(piperidin-4-yl)methoxy]pyridin-4-yl}-1H-pyrazol-1-yl)phenyl]piperazine, Maternal embryonic leucine zipper kinase | | Authors: | Sprague, E.R, Puleo, D.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
5IH8
 
 | | MELK in complex with NVS-MELK1 | | Descriptor: | Maternal embryonic leucine zipper kinase, N-[(pyridin-2-yl)methyl]-4-[4-(pyridin-4-yl)-1H-pyrazol-1-yl]benzamide | | Authors: | Sprague, E.R, Puleo, D.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
1UN5
 
 | | ARH-II, AN ANGIOGENIN/RNASE A CHIMERA | | Descriptor: | ANGIOGENIN, CITRIC ACID | | Authors: | Holloway, D.E, Baker, M.D, Acharya, K.R. | | Deposit date: | 2003-09-04 | | Release date: | 2004-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Studies on Structural Features that Determine the Enzymatic Specificity and Potency of Human Angiogenin: Thr44, Thr80 and Residues 38-41
Biochemistry, 43, 2004
|
|
5J7J
 
 | |
4YJ2
 
 | | Crystal structure of tubulin bound to MI-181 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,6-dimethyl-2-[(E)-2-(pyridin-3-yl)ethenyl]-1,3-benzothiazole, CALCIUM ION, ... | | Authors: | McNamara, D.E, Torres, J.Z, Yeates, T.O. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of potent anticancer compounds bound to tubulin.
Protein Sci., 24, 2015
|
|
1UN4
 
 | |
5I1S
 
 | | Villin headpiece subdomain with a Lys30 to APC substitution | | Descriptor: | D-Villin headpiece subdomain, Villin-1 | | Authors: | Kreitler, D.F, Mortenson, D.E, Gellman, S.H, Forest, K.T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Effects of Single alpha-to-beta Residue Replacements on Structure and Stability in a Small Protein: Insights from Quasiracemic Crystallization.
J.Am.Chem.Soc., 138, 2016
|
|
1W17
 
 | |
1UN3
 
 | |
6REM
 
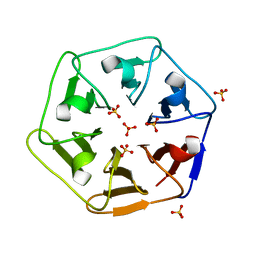 | | Crystal structure of Pizza6-SH with Sulphate ion | | Descriptor: | Pizza6-SH, SULFATE ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Artificial beta-Propeller Protein-based Hydrolases
To Be Published
|
|
4X7I
 
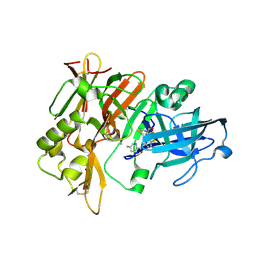 | | Crystal Structure of BACE with amino thiazine inhibitor LY2886721 | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{3-[(4aS,7aS)-2-amino-4a,5-dihydro-4H-furo[3,4-d][1,3]thiazin-7a(7H)-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Authors: | Timm, D.E. | | Deposit date: | 2014-12-09 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Potent BACE1 Inhibitor LY2886721 Elicits Robust Central A beta Pharmacodynamic Responses in Mice, Dogs, and Humans.
J.Neurosci., 35, 2015
|
|
1W1A
 
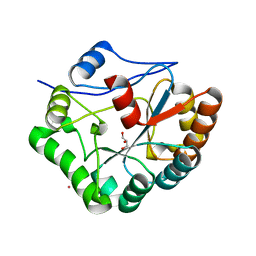 | | Structure of Bacillus subtilis PdaA in complex with NAG, a family 4 Carbohydrate esterase. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, CADMIUM ION, GLYCEROL, ... | | Authors: | Blair, D.E, van Aalten, D.M.F. | | Deposit date: | 2004-06-18 | | Release date: | 2005-01-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine.
FEBS Lett., 570, 2004
|
|
4YBI
 
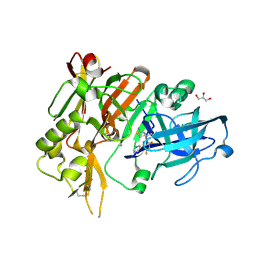 | | Crystal structure of BACE with amino thiazine inhibitor LY2811376 | | Descriptor: | (4S)-4-[2,4-difluoro-5-(pyrimidin-5-yl)phenyl]-4-methyl-5,6-dihydro-4H-1,3-thiazin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Timm, D.E. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Robust central reduction of amyloid-beta in humans with an orally available, non-peptidic beta-secretase inhibitor.
J.Neurosci., 31, 2011
|
|
4YJ3
 
 | | Crystal structure of tubulin bound to compound 2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(4-ethoxyphenyl)-3-(2-methoxyphenyl)-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazine, CALCIUM ION, ... | | Authors: | McNamara, D.E, Torres, J.Z, Yeates, T.O. | | Deposit date: | 2015-03-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structures of potent anticancer compounds bound to tubulin.
Protein Sci., 24, 2015
|
|
1W49
 
 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP and Mg | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
5I1N
 
 | | Villin headpiece subdomain with a Gln26 to beta-3-homoglutamine substitution | | Descriptor: | D-Villin headpiece subdomain, Villin-1 | | Authors: | Kreitler, D.F, Mortenson, D.E, Gellman, S.H, Forest, K.T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Effects of Single alpha-to-beta Residue Replacements on Structure and Stability in a Small Protein: Insights from Quasiracemic Crystallization.
J.Am.Chem.Soc., 138, 2016
|
|
1W4B
 
 | | P4 protein from PHI12 in complex with product (AMPcPP Mg 22C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
