3VW7
 
 | | Crystal structure of human protease-activated receptor 1 (PAR1) bound with antagonist vorapaxar at 2.2 angstrom | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Proteinase-activated receptor 1, ... | | Authors: | Zhang, C, Srinivasan, Y, Arlow, D.H, Fung, J.J, Palmer, D, Zheng, Y, Green, H.F, Pandey, A, Dror, R.O, Shaw, D.E, Weis, W.I, Coughlin, S.R, Kobilka, B.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structure of human protease-activated receptor 1
Nature, 492, 2012
|
|
8F36
 
 | |
8F37
 
 | |
2C1G
 
 | | Structure of Streptococcus pneumoniae peptidoglycan deacetylase (SpPgdA) | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, PEPTIDOGLYCAN GLCNAC DEACETYLASE, ... | | Authors: | Blair, D.E, Schuttelkopf, A.W, MacRae, J.I, van Aalten, D.M.F. | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
2C76
 
 | | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins | | Descriptor: | AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE, N-PROPARGYL-1(S)-AMINOINDAN | | Authors: | Li, M, Binda, C, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins
Biochemistry, 45, 2006
|
|
2C73
 
 | | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins | | Descriptor: | AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE, N-PROPARGYL-1(S)-AMINOINDAN | | Authors: | Li, M, Binda, C, Mattevi, A, Edmondson, D.E. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional Role of the Aromatic Cage in Human Monoamine Oxidase B: Structures and Catalytic Properties of Tyr435 Mutant Proteins
Biochemistry, 45, 2006
|
|
8ESF
 
 | |
2C01
 
 | | Crystal Structures of Eosinophil-derived Neurotoxin in Complex with the Inhibitors 5'-ATP, Ap3A, Ap4A and Ap5A | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, NONSECRETORY RIBONUCLEASE | | Authors: | Baker, M.D, Holloway, D.E, Swaminathan, G.J, Acharya, K.R. | | Deposit date: | 2005-08-24 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal Structures of Eosinophil-Derived Neurotoxin (Edn) in Complex with the Inhibitors 5'- ATP, Ap(3)A, Ap(4)A, and Ap(5)A.
Biochemistry, 45, 2006
|
|
2BK4
 
 | | Human Monoamine Oxidase B: I199F mutant in complex with rasagiline | | Descriptor: | (1R)-N-(prop-2-en-1-yl)-2,3-dihydro-1H-inden-1-amine, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Edmondson, D.E, Mattevi, A, Hubalek, F, Khalil, A, Li, M, Castagnoli, N. | | Deposit date: | 2005-02-10 | | Release date: | 2005-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Demonstration of Isoleucine 199 as a Structural Determinant for the Selective Inhibition of Human Monoamine Oxidase B by Specific Reversible Inhibitors
J.Biol.Chem., 280, 2005
|
|
2BK3
 
 | | Human Monoamine Oxidase B in complex with Farnesol | | Descriptor: | (2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-02-10 | | Release date: | 2005-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Demonstration of isoleucine 199 as a structural determinant for the selective inhibition of human monoamine oxidase B by specific reversible inhibitors.
J. Biol. Chem., 280, 2005
|
|
2BWL
 
 | | Murine angiogenin, phosphate complex | | Descriptor: | ANGIOGENIN, PHOSPHATE ION | | Authors: | Holloway, D.E, Chavali, G.B, Hares, M.C, Subramanian, V, Acharya, K.R. | | Deposit date: | 2005-07-15 | | Release date: | 2005-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of Murine Angiogenin: Features of the Substrate- and Cell-Binding Regions and Prospects for Inhibitor-Binding Studies.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A2P
 
 | | Solution structure of SelM from Mus musculus | | Descriptor: | Selenoprotein M | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
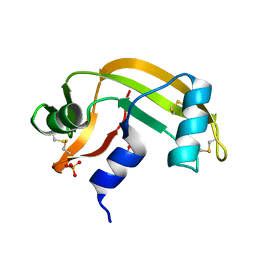
2AET
 
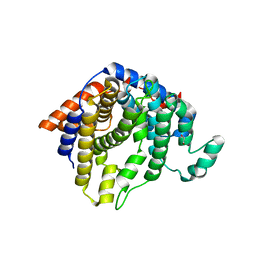 | | R304K trichodiene synthase: Complex with Mg, pyrophosphate, and (4S)-7-azabisabolene | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-07-23 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Role of arginine-304 in the diphosphate-triggered active site closure mechanism of trichodiene synthase.
Biochemistry, 44, 2005
|
|
2AQP
 
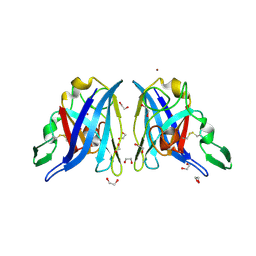 | | CU/ZN superoxide dismutase from neisseria meningitidis E73A mutant | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | DiDonato, M, Kassmann, C.J, Bruns, C.K, Cabelli, D.E, Cao, Z, Tabatabai, L.B, Kroll, J.S, Getzoff, E.D. | | Deposit date: | 2005-08-18 | | Release date: | 2006-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | CU/ZN superoxide dismutase from neisseria meningitidis E73A mutant
To be Published
|
|
3TJW
 
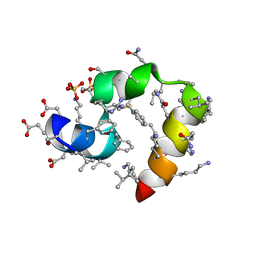 | | Crystal Structure of Quasiracemic Villin Headpiece Subdomain Containing (F5Phe10) Substitution | | Descriptor: | D-Villin-1, L-Villin-1, SULFATE ION | | Authors: | Mortenson, D.E, Satyshur, K.A, Gellman, S.H, Forest, K.T. | | Deposit date: | 2011-08-25 | | Release date: | 2012-01-25 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Quasiracemic crystallization as a tool to assess the accommodation of noncanonical residues in nativelike protein conformations.
J.Am.Chem.Soc., 134, 2012
|
|
2AGI
 
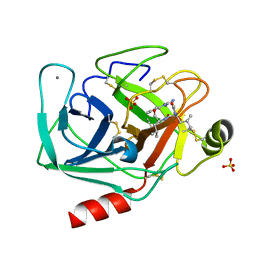 | | The leupeptin-trypsin covalent complex at 1.14 A resolution | | Descriptor: | CALCIUM ION, SULFATE ION, beta-trypsin, ... | | Authors: | Radisky, E.S, Lee, J.M, Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
8FZY
 
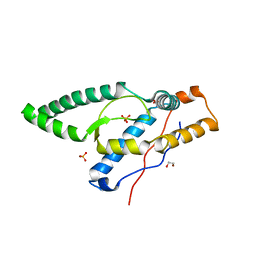 | |
2AHQ
 
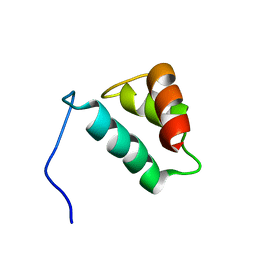 | |
8FZZ
 
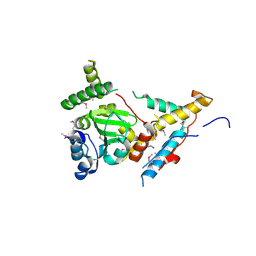 | |
8G0K
 
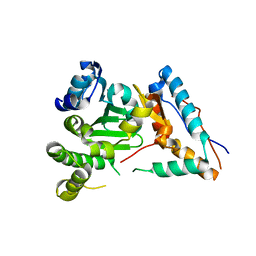 | |
2C66
 
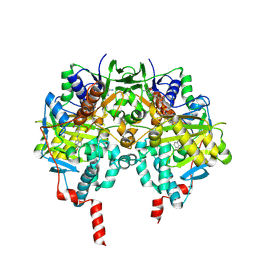 | | MAO inhibition by rasagiline analogues | | Descriptor: | 4-HYDROXY-N-PROPARGYL-1(R)-AMINOINDAN, AMINE OXIDASE (FLAVIN-CONTAINING) B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-11-07 | | Release date: | 2006-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding of Rasagiline-Related Inhibitors to Human Monoamine Oxidases: A Kinetic and Crystallographic Analysis.
J.Med.Chem., 48, 2005
|
|
2E34
 
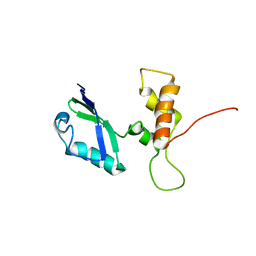 | | L11 structure with RDC and RG refinement | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Markus, M.A, Choli-Papadopoulous, T, Schwieters, C.D, Krueger, S, Draper, D.E, Wang, Y.X. | | Deposit date: | 2006-11-20 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of free L11 and functional dynamics of L11 in free, L11-rRNA(58 nt) binary and L11-rRNA(58 nt)-thiostrepton ternary complexes
J.Mol.Biol., 367, 2007
|
|
2E4O
 
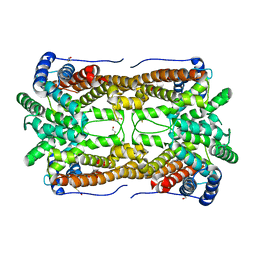 | | X-ray Crystal Structure of Aristolochene Synthase from Aspergillus terreus and the Evolution of Templates for the Cyclization of Farnesyl Diphosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Di Costanzo, L, Cane, D.E, Christianson, D.W. | | Deposit date: | 2006-12-15 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate.
Biochemistry, 46, 2007
|
|
7S9Z
 
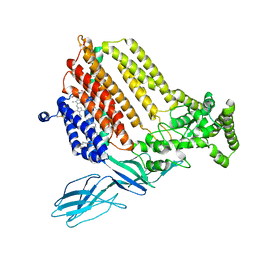 | | Helicobacter Hepaticus CcsBA Closed Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
7S9Y
 
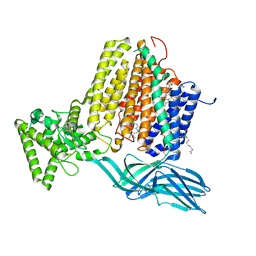 | | Helicobacter Hepaticus CcsBA Open Conformation | | Descriptor: | Cytochrome c biogenesis protein, HEME B/C, PHOSPHATIDYLETHANOLAMINE | | Authors: | Mendez, D.L, Lowder, E.P, Tillman, D.E, Sutherland, M.C, Collier, A.L, Rau, M.J, Fitzpatrick, J.A, Kranz, R.G. | | Deposit date: | 2021-09-21 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM of CcsBA reveals the basis for cytochrome c biogenesis and heme transport.
Nat.Chem.Biol., 18, 2022
|
|
