1GV7
 
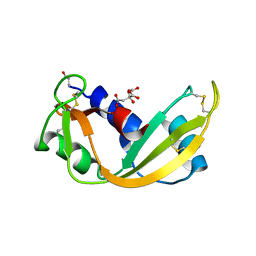 | | ARH-I, an angiogenin/RNase A chimera | | Descriptor: | ANGIOGENIN, CITRIC ACID | | Authors: | Holloway, D.E, Shapiro, R, Hares, M.C, Leonidas, D.D, Acharya, K.R. | | Deposit date: | 2002-02-06 | | Release date: | 2002-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Guest-Host Crosstalk in an Angiogenin-RNase A Chimeric Protein
Biochemistry, 41, 2002
|
|
1FEO
 
 | |
1FJC
 
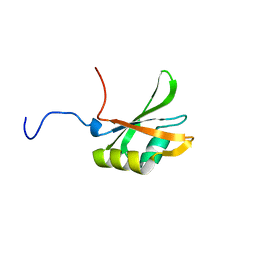 | |
1I52
 
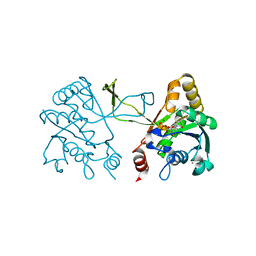 | | CRYSTAL STRUCTURE OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHASE, CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, M, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-02-23 | | Release date: | 2001-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1GOS
 
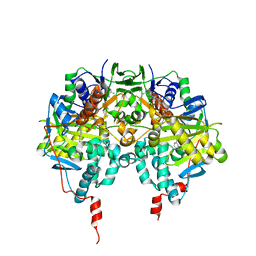 | | Human Monoamine Oxidase B | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MONOAMINE OXIDASE, N-[(E)-METHYL](PHENYL)-N-[(E)-2-PROPENYLIDENE]METHANAMINIUM | | Authors: | Binda, C, Newton-Vinson, P, Hubalek, F, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2001-10-26 | | Release date: | 2001-11-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Human Monoamine Oxidase B, a Drug Target for the Treatment of Neurological Disorders
Nat.Struct.Biol., 9, 2001
|
|
1GQV
 
 | | Atomic Resolution (0.98A) Structure of Eosinophil-Derived Neurotoxin | | Descriptor: | ACETATE ION, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Swaminathan, G.J, Holloway, D.E, Veluraja, K, Acharya, K.R. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution (0.98 A) Structure of Eosinophil-Derived Neurotoxin
Biochemistry, 41, 2002
|
|
1IG0
 
 | | Crystal Structure of yeast Thiamin Pyrophosphokinase | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Thiamin pyrophosphokinase | | Authors: | Baker, L.-J, Dorocke, J.A, Harris, R.A, Timm, D.E. | | Deposit date: | 2001-04-16 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of yeast thiamin pyrophosphokinase.
Structure, 9, 2001
|
|
1INJ
 
 | | CRYSTAL STRUCTURE OF THE APO FORM OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHETASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHETASE, CALCIUM ION | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, A, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-05-14 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1HQX
 
 | | R308K ARGINASE VARIANT | | Descriptor: | ARGINASE, MANGANESE (II) ION | | Authors: | Lavulo, L.T, Sossong Jr, T.M, Brigham-Burke, M.R, Doyle, M.L, Cox, J.D, Christianson, D.W, Ash, D.E. | | Deposit date: | 2000-12-20 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Subunit-subunit interactions in trimeric arginase. Generation of active monomers by mutation of a single amino acid.
J.Biol.Chem., 276, 2001
|
|
1HQ5
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH S-(2-BORONOETHYL)-L-CYSTEINE, AN L-ARGININE ANALOGUE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, S-2-(BORONOETHYL)-L-CYSTEINE | | Authors: | Kim, N.N, Cox, J.D, Baggio, R.F, Emig, F.A, Mistry, S.K, Harper, S.L, Speicher, D.W, Morris Jr, S.M, Ash, D.E, Traish, A, Christianson, D.W. | | Deposit date: | 2000-12-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing erectile function: S-(2-boronoethyl)-L-cysteine binds to arginase as a transition state analogue and enhances smooth muscle relaxation in human penile corpus cavernosum.
Biochemistry, 40, 2001
|
|
1HQH
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH NOR-N-HYDROXY-L-ARGININE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, NOR-N-OMEGA-HYDROXY-L-ARGININE | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
1FJ7
 
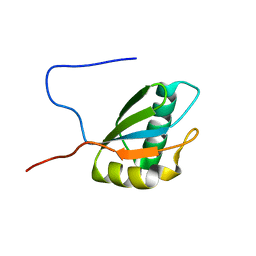 | |
1HM7
 
 | |
1HQF
 
 | | CRYSTAL STRUCTURE OF THE BINUCLEAR MANGANESE METALLOENZYME ARGINASE COMPLEXED WITH N-HYDROXY-L-ARGININE | | Descriptor: | ARGINASE 1, MANGANESE (II) ION, N-OMEGA-HYDROXY-L-ARGININE | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
1HM4
 
 | |
1HQG
 
 | | CRYSTAL STRUCTURE OF THE H141C ARGINASE VARIANT COMPLEXED WITH PRODUCTS ORNITHINE AND UREA | | Descriptor: | ARGINASE 1, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
1IM1
 
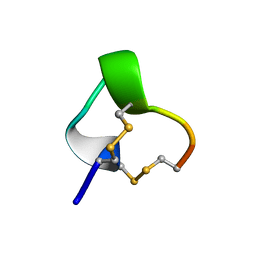 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN IM1, 20 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN IM1 | | Authors: | Rogers, J.P, Luginbuhl, P, Shen, G.S, Mccabe, R.T, Stevens, R.C, Wemmer, D.E. | | Deposit date: | 1998-11-18 | | Release date: | 1999-06-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of alpha-conotoxin ImI and comparison to other conotoxins specific for neuronal nicotinic acetylcholine receptors.
Biochemistry, 38, 1999
|
|
1FVT
 
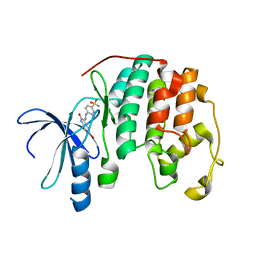 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(2Z)-2-(5-bromo-2-oxo-1,2-dihydro-3H-indol-3-ylidene)hydrazinyl]benzene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
1FYY
 
 | | HPRT GENE MUTATION HOTSPOT WITH A BPDE2(10R) ADDUCT | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*TP*GP*CP*CP*CP*TP*TP*GP*AP*CP*TP*A)-3', HPRT DNA WITH BENZO[A]PYRENE-ADDUCTED DA7 | | Authors: | Volk, D.E, Rice, J.S, Luxon, B.A, Yeh, H.J.C, Liang, C, Xie, G, Sayer, J.M, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2000-10-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR evidence for syn-anti interconversion of a trans opened (10R)-dA adduct of benzo[a]pyrene (7S,8R)-diol (9R,10S)-epoxide in a DNA duplex.
Biochemistry, 39, 2000
|
|
1INI
 
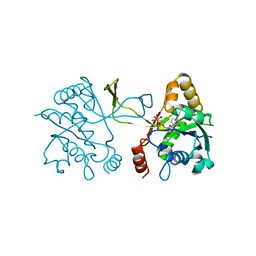 | | CRYSTAL STRUCTURE OF 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL (CDP-ME) SYNTHETASE (YGBP) INVOLVED IN MEVALONATE INDEPENDENT ISOPRENOID BIOSYNTHESIS, COMPLEXED WITH CDP-ME AND MG2+ | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, 4-DIPHOSPHOCYTIDYL-2-C-METHYLERYTHRITOL SYNTHETASE, MAGNESIUM ION | | Authors: | Richard, S.B, Bowman, M.E, Kwiatkowski, W, Kang, I, Chow, C, Lillo, A, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-05-14 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of 4-diphosphocytidyl-2-C- methylerythritol synthetase involved in mevalonate- independent isoprenoid biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1FOY
 
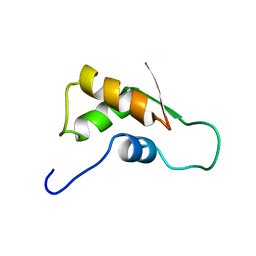 | | THE RNA BINDING DOMAIN OF RIBOSOMAL PROTEIN L11: THREE-DIMENSIONAL STRUCTURE OF THE RNA-BOUND FORM OF THE PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RIBOSOMAL PROTEIN L11 | | Authors: | Hinck, A.P, Markus, M.A, Huang, S, Grzesiek, S, Kustanovich, I, Draper, D.E, Torchia, D.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-11-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The RNA binding domain of ribosomal protein L11: three-dimensional structure of the RNA-bound form of the protein and its interaction with 23 S rRNA.
J.Mol.Biol., 274, 1997
|
|
1GMY
 
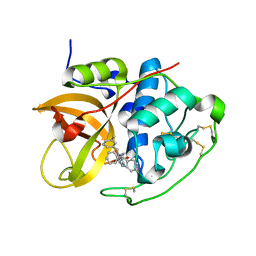 | | Cathepsin B complexed with dipeptidyl nitrile inhibitor | | Descriptor: | 2-AMINOETHANIMIDIC ACID, 3-METHYLPHENYLALANINE, CATHEPSIN B, ... | | Authors: | Greenspan, P.D, Clark, K.L, Tommasi, R.A, Cowen, S.D, McQuire, L.W, Farley, D.L, van Duzer, J.H, Goldberg, R.L, Zhou, H, Du, Z, Fitt, J.J, Coppa, D.E, Fang, Z, Macchia, W, Zhu, L, Capparelli, M.P, Goldstein, R, Wigg, A.M, Doughty, J.R, Bohacek, R.S, Knap, A.K. | | Deposit date: | 2001-09-25 | | Release date: | 2002-09-19 | | Last modified: | 2017-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of Dipeptidyl Nitriles as Potent and Selective Inhibitors of Cathepsin B Through Structure-Based Drug Design
J.Med.Chem., 44, 2001
|
|
1J70
 
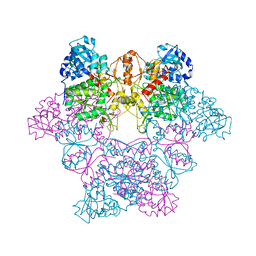 | | CRYSTAL STRUCTURE OF YEAST ATP SULFURYLASE | | Descriptor: | ATP SULPHURYLASE, PHOSPHATE ION, SODIUM ION | | Authors: | Lalor, D.J, Schnyder, T, Saridakis, V, Pilloff, D.E, Dong, A, Tang, H, Leyh, T.S, Pai, E.F. | | Deposit date: | 2001-05-15 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of a truncated form of Saccharomyces cerevisiae ATP sulfurylase: C-terminal domain essential for oligomer formation but not for activity.
Protein Eng., 16, 2003
|
|
1HYO
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH 4-(HYDROXYMETHYLPHOSPHINOYL)-3-OXO-BUTANOIC ACID | | Descriptor: | 4-[HYDROXY-[METHYL-PHOSPHINOYL]]-3-OXO-BUTANOIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Bateman, R.L, Bhanumoorthy, P, Witte, J.F, McClard, R.W, Grompe, M, Timm, D.E. | | Deposit date: | 2001-01-21 | | Release date: | 2001-02-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanistic inferences from the crystal structure of fumarylacetoacetate hydrolase with a bound phosphorus-based inhibitor.
J.Biol.Chem., 276, 2001
|
|
1HZ5
 
 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS, WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
