1AJR
 
 | | REFINEMENT AND COMPARISON OF THE CRYSTAL STRUCTURES OF PIG CYTOSOLIC ASPARTATE AMINOTRANSFERASE AND ITS COMPLEX WITH 2-METHYLASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE | | Authors: | Rhee, S, Silva, M.M, Hyde, C.C, Rogers, P.H, Metzler, C.M, Metzler, D.E, Arnone, A. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate.
J.Biol.Chem., 272, 1997
|
|
1A2G
 
 | |
1A2F
 
 | |
1BBH
 
 | |
1BBI
 
 | |
1ASH
 
 | | THE STRUCTURE OF ASCARIS HEMOGLOBIN DOMAIN I AT 2.2 ANGSTROMS RESOLUTION: MOLECULAR FEATURES OF OXYGEN AVIDITY | | Descriptor: | HEMOGLOBIN (OXY), OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, J, Mathews, F.S, Kloek, A.P, Goldberg, D.E. | | Deposit date: | 1995-01-06 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of Ascaris hemoglobin domain I at 2.2 A resolution: molecular features of oxygen avidity.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1TLP
 
 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | Descriptor: | CALCIUM ION, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, THERMOLYSIN, ... | | Authors: | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
1AJS
 
 | | REFINEMENT AND COMPARISON OF THE CRYSTAL STRUCTURES OF PIG CYTOSOLIC ASPARTATE AMINOTRANSFERASE AND ITS COMPLEX WITH 2-METHYLASPARTATE | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, ASPARTATE AMINOTRANSFERASE | | Authors: | Rhee, S, Silva, M.M, Hyde, C.C, Rogers, P.H, Metzler, C.M, Metzler, D.E, Arnone, A. | | Deposit date: | 1997-05-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate.
J.Biol.Chem., 272, 1997
|
|
1C05
 
 | | SOLUTION STRUCTURE OF RIBOSOMAL PROTEIN S4 DELTA 41, REFINED WITH DIPOLAR COUPLINGS (MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | RIBOSOMAL PROTEIN S4 DELTA 41 | | Authors: | Markus, M.A, Gerstner, R.B, Draper, D.E, Torchia, D.A. | | Deposit date: | 1999-07-14 | | Release date: | 1999-09-29 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Refining the overall structure and subdomain orientation of ribosomal protein S4 delta41 with dipolar couplings measured by NMR in uniaxial liquid crystalline phases.
J.Mol.Biol., 292, 1999
|
|
5VTJ
 
 | | Structure of Pin1 WW Domain Sequence 1 Substituted with [S,S]ACPC | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | Authors: | Mortenson, D.E, Kreitler, D.F, Thomas, N.C, Gellman, S.H, Forest, K.T. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evaluation of beta-Amino Acid Replacements in Protein Loops: Effects on Conformational Stability and Structure.
Chembiochem, 19, 2018
|
|
5VTI
 
 | | Structure of Pin1 WW Domain Sequence 3 with [R,R]-ACPC Loop Substitution | | Descriptor: | CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Kreitler, D.F, Thomas, N.C, Gellman, S.H, Forest, K.T. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluation of beta-Amino Acid Replacements in Protein Loops: Effects on Conformational Stability and Structure.
Chembiochem, 19, 2018
|
|
4V41
 
 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
4V4R
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-09-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.9 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V7K
 
 | | Structure of RelE nuclease bound to the 70S ribosome (postcleavage state) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | Deposit date: | 2009-11-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
4V4S
 
 | | Crystal structure of the whole ribosomal complex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.76 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
4V4T
 
 | | Crystal structure of the whole ribosomal complex with a stop codon in the A-site. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Petry, S, Brodersen, D.E, Murphy IV, F.V, Dunham, C.M, Selmer, M, Tarry, M.J, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2005-10-12 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.46 Å) | | Cite: | Crystal Structures of the Ribosome in Complex with Release Factors RF1 and RF2 Bound to a Cognate Stop Codon.
Cell(Cambridge,Mass.), 123, 2005
|
|
5WW0
 
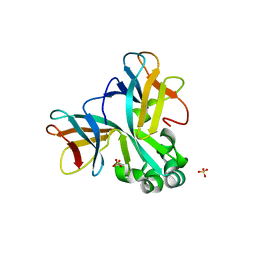 | | Crystal structure of Set7, a novel histone methyltransferase in Schizossacharomyces pombe | | Descriptor: | SET domain-containing protein 7, SULFATE ION | | Authors: | Mevius, D.E.H.F, Shen, Y, Morishita, M, Carrozzini, B, Caliandro, R, di Luccio, E. | | Deposit date: | 2016-12-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Set7 Is a H3K37 Methyltransferase in Schizosaccharomyces pombe and Is Required for Proper Gametogenesis.
Structure, 27, 2019
|
|
5W87
 
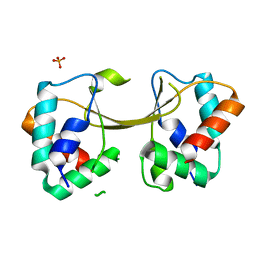 | |
5VTK
 
 | | Structure of Pin1 WW Domain Variant 1 with beta3-Ser Loop Substitution | | Descriptor: | CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Mortenson, D.E, Kreitler, D.F, Thomas, N.C, Gellman, S.H, Forest, K.T. | | Deposit date: | 2017-05-17 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Evaluation of beta-Amino Acid Replacements in Protein Loops: Effects on Conformational Stability and Structure.
Chembiochem, 19, 2018
|
|
6GFM
 
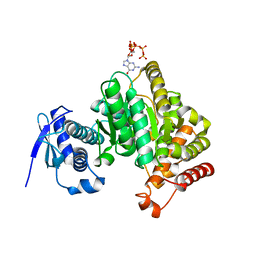 | | Crystal structure of the Escherichia coli nucleosidase PpnN (pppGpp-form) | | Descriptor: | Pyrimidine/purine nucleotide 5'-monophosphate nucleosidase, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Zhang, Y, Baerentsen, R.L, Gerdes, K, Brodersen, D.E. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | (p)ppGpp Regulates a Bacterial Nucleosidase by an Allosteric Two-Domain Switch.
Mol.Cell, 74, 2019
|
|
6GFL
 
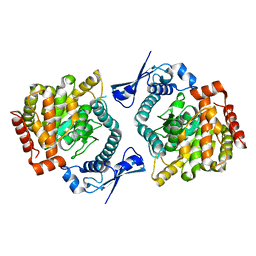 | |
6HPB
 
 | |
1F4V
 
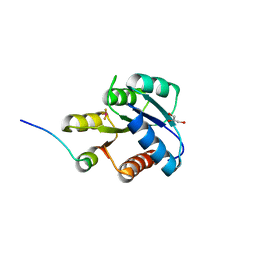 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-06-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
1FOX
 
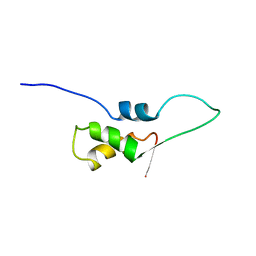 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, 33 STRUCTURES | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
1FOW
 
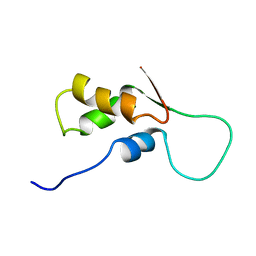 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
