5W87
 
 | |
5ZX5
 
 | | 3.3 angstrom structure of mouse TRPM7 with EDTA | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 7 | | Authors: | Zhang, J, Li, Z, Duan, J, Li, J, Hulse, R.E, Santa-Cruz, A, Abiria, S.A, Krapivinsky, G, Clapham, D.E. | | Deposit date: | 2018-05-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1VLS
 
 | | LIGAND BINDING DOMAIN OF THE WILD-TYPE ASPARTATE RECEPTOR | | Descriptor: | ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Yeh, J.I, Biemann, H.-P, Prive, G, Pandit, J, Koshland Junior, D.E. | | Deposit date: | 1996-09-17 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor.
J.Mol.Biol., 262, 1996
|
|
1VLT
 
 | | LIGAND BINDING DOMAIN OF THE WILD-TYPE ASPARTATE RECEPTOR WITH ASPARTATE | | Descriptor: | ASPARTATE RECEPTOR, ASPARTIC ACID | | Authors: | Kim, S.-H, Yeh, J.I, Biemann, H.-P, Prive, G, Pandit, J, Koshland Junior, D.E. | | Deposit date: | 1996-09-17 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor.
J.Mol.Biol., 262, 1996
|
|
1BOF
 
 | | GI-ALPHA-1 BOUND TO GDP AND MAGNESIUM | | Descriptor: | GI ALPHA 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Coleman, D.E, Sprang, S.R. | | Deposit date: | 1998-08-04 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the G protein Gi alpha 1 complexed with GDP and Mg2+: a crystallographic titration experiment.
Biochemistry, 37, 1998
|
|
6DU8
 
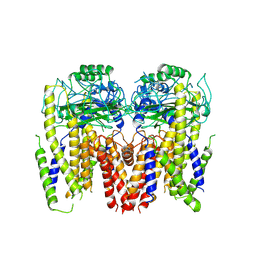 | | Human Polycsytin 2-l1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystic kidney disease 2-like 1 protein | | Authors: | Hulse, R.E, Clapham, D.E, Li, Z, Huang, R.K, Zhang, J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Cryo-EM structure of the polycystin 2-l1 ion channel.
Elife, 7, 2018
|
|
5ED0
 
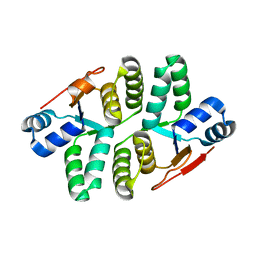 | |
5DZ2
 
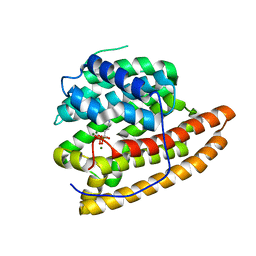 | | Geosmin synthase from Streptomyces coelicolor N-terminal domain complexed with three Mg2+ ions and alendronic acid | | Descriptor: | ALENDRONATE, Germacradienol/geosmin synthase, MAGNESIUM ION | | Authors: | Harris, G.G, Lombardi, P.M, Pemberton, T.A, Matsui, T, Weiss, T.M, Cole, K.E, Koksal, M, Murphy, F.V, Vedula, L.S, Chou, W.K.W, Cane, D.E, Christianson, D.W. | | Deposit date: | 2015-09-25 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Structural Studies of Geosmin Synthase, a Bifunctional Sesquiterpene Synthase with alpha alpha Domain Architecture That Catalyzes a Unique Cyclization-Fragmentation Reaction Sequence.
Biochemistry, 54, 2015
|
|
5ECY
 
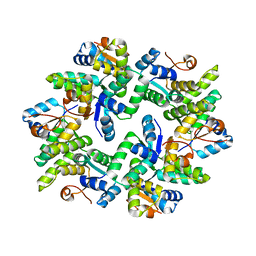 | |
5ECW
 
 | |
2QS4
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
6DRH
 
 | | ADP-ribosyltransferase toxin/immunity pair | | Descriptor: | ADP-ribosyl-(Dinitrogen reductase) hydrolase, MAGNESIUM ION, PAAR repeat-containing protein, ... | | Authors: | Bosch, D.E, Ting, S, Allaire, M, Mougous, J.D. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins.
Cell, 175, 2018
|
|
1EEM
 
 | | GLUTATHIONE TRANSFERASE FROM HOMO SAPIENS | | Descriptor: | GLUTATHIONE, GLUTATHIONE-S-TRANSFERASE, SULFATE ION | | Authors: | Board, P, Coggan, M, Chelvanayagam, G, Easteal, S, Jermiin, L.S, Schulte, G.K, Danley, D.E, Hoth, L.R, Griffor, M.C, Kamath, A.V, Rosner, M.H, Chrunyk, B.A, Perregaux, D.E, Gabel, C.A, Geoghegan, K.F, Pandit, J. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification, characterization, and crystal structure of the Omega class glutathione transferases.
J.Biol.Chem., 275, 2000
|
|
6ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
2QS1
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-4,5-dibromothiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QTX
 
 | | Crystal structure of an Hfq-like protein from Methanococcus jannaschii | | Descriptor: | Uncharacterized protein MJ1435 | | Authors: | Nielsen, J.S, Boggild, A, Andersen, C.B.F, Nielsen, G, Boysen, A, Brodersen, D.E, Valentin-Hansen, P. | | Deposit date: | 2007-08-03 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Hfq-like protein in archaea: Crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii.
Rna, 13, 2007
|
|
6DRE
 
 | | ADP-ribosyltransferase toxin/immunity pair | | Descriptor: | ADP-ribosyl-(Dinitrogen reductase) hydrolase, MAGNESIUM ION, PAAR repeat-containing protein | | Authors: | Bosch, D.E, Ting, S, Allaire, M, Mougous, J.D. | | Deposit date: | 2018-06-11 | | Release date: | 2018-10-31 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins.
Cell, 175, 2018
|
|
6DRW
 
 | |
2Q9Z
 
 | | Trichodiene synthase: Complex with inorganic pyrophosphate resulting from the reaction with 2-fluorofarnesyl diphosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Zhao, Y, Coates, R.M, Koyama, T, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-06-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Exploring biosynthetic diversity with trichodiene synthase.
Arch.Biochem.Biophys., 466, 2007
|
|
1RYC
 
 | | CYTOCHROME C PEROXIDASE W191G FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | BENZIMIDAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Musah, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
6HPB
 
 | |
6GW6
 
 | |
6HPC
 
 | |
1ZDM
 
 | | Crystal Structure of Activated CheY Bound to Xe | | Descriptor: | Chemotaxis protein cheY, MANGANESE (II) ION, XENON | | Authors: | Lowery, T.J, Doucleff, M, Ruiz, E.J, Rubin, S.M, Pines, A, Wemmer, D.E. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR.
Protein Sci., 14, 2005
|
|
6ZN8
 
 | | Crystal structure of the H. influenzae VapXD toxin-antitoxin complex | | Descriptor: | Endoribonuclease VapD, VapX | | Authors: | Bertelsen, M.B, Senissar, M, Nielsen, M.H, Bisiak, F, Cunha, M.V, Molinaro, A.L, Daines, D.A, Brodersen, D.E. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Structural Basis for Toxin Inhibition in the VapXD Toxin-Antitoxin System.
Structure, 29, 2021
|
|
