6PFH
 
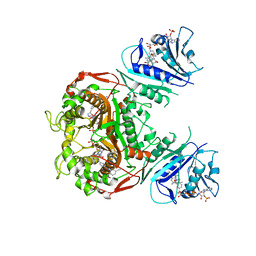 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-cyanophenyl)acetic acid. | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, ... | | Authors: | Czyzyk, D.C, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
7JJU
 
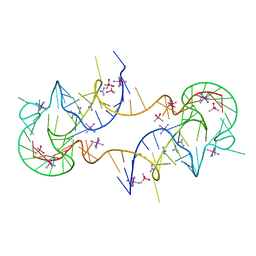 | | Crystal structure of en exoribonuclease-resistant RNA (xrRNA) from Potato leafroll virus (PLRV) | | Descriptor: | CACODYLATE ION, Guanidinium, IRIDIUM HEXAMMINE ION, ... | | Authors: | Steckelberg, A.-L, Vicens, Q, Auffinger, P, Costantino, D.C, Nix, J.C, Kieft, J.S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The crystal structure of a Polerovirus exoribonuclease-resistant RNA shows how diverse sequences are integrated into a conserved fold.
Rna, 26, 2020
|
|
7JMN
 
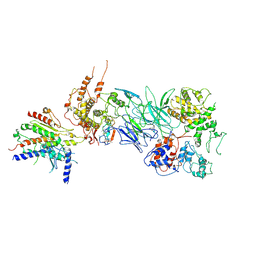 | | Tail module of Mediator complex | | Descriptor: | MED15, Mediator of RNA polymerase II transcription subunit 14, Mediator of RNA polymerase II transcription subunit 16, ... | | Authors: | Zhang, H.Q, Chen, D.C. | | Deposit date: | 2020-08-02 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
2FG8
 
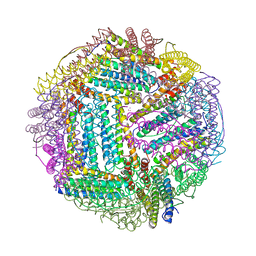 | | Structure of Human Ferritin L Chain | | Descriptor: | CESIUM ION, Ferritin light chain | | Authors: | Wang, Z.M, Li, C, Ellenburg, M.P, Ruble, J.R, Ho, J.X, Carter, D.C. | | Deposit date: | 2005-12-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of human ferritin L chain.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2HXW
 
 | | Crystal Structure of Peb3 from Campylobacter jejuni | | Descriptor: | CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Rangarajan, E.S, Bhatia, S, Watson, D.C, Munger, C, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-04 | | Release date: | 2007-05-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural context for protein N-glycosylation in bacteria: The structure of PEB3, an adhesin from Campylobacter jejuni.
Protein Sci., 16, 2007
|
|
2FG4
 
 | | Structure of Human Ferritin L Chain | | Descriptor: | CADMIUM ION, Ferritin light chain | | Authors: | Wang, Z, Li, C, Ellenburg, M, Ruble, J, Ho, J.X, Carter, D.C. | | Deposit date: | 2005-12-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ferritin L chain.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3T8V
 
 | | A bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases | | Descriptor: | M1 family aminopeptidase, MAGNESIUM ION, N-[(2-{2-[(N-{(2S,3R)-3-amino-4-[4-(benzyloxy)phenyl]-2-hydroxybutanoyl}-L-alanyl)amino]ethoxy}ethoxy)acetyl]-4-benzoyl-L-phenylalanyl-N~6~-hex-5-ynoyllysinamide, ... | | Authors: | McGowan, S, Klemba, M, Greebaum, D.C. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2FQA
 
 | | Violacin A | | Descriptor: | violacin 1 | | Authors: | Ireland, D.C, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-01-17 | | Release date: | 2006-01-31 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Discovery and Characterization of a Linear Cyclotide from Viola odorata: Implications for the Processing of Circular Proteins
J.Mol.Biol., 357, 2006
|
|
2HHN
 
 | | Cathepsin S in complex with non covalent arylaminoethyl amide. | | Descriptor: | Cathepsin S, N-[(1R)-1-[(BENZYLSULFONYL)METHYL]-2-{[(1S)-1-METHYL-2-{[4-(TRIFLUOROMETHOXY)PHENYL]AMINO}ETHYL]AMINO}-2-OXOETHYL]MORPHOLINE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2006-06-28 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis and SAR of arylaminoethyl amides as noncovalent inhibitors of cathepsin S: P3 cyclic ethers
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FFX
 
 | | Structure of Human Ferritin L. Chain | | Descriptor: | CADMIUM ION, SULFATE ION, ferritin light chain | | Authors: | Wang, Z.M, Li, C, Ellenburg, M.P, Soitsman, E.M, Ruble, J.R, Wright, B.S, Ho, J.X, Carter, D.C. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human ferritin L chain
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
3T8W
 
 | | A bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, N-((2R,3S,6S,18S,21S)-2-amino-18-(4-benzoylbenzyl)-21-carbamoyl-3-hydroxy-6-(naphthalen-2-ylmethyl)-4,7,16,19-tetraoxo-1-phenyl-11,14-dioxa-5,8,17,20-tetraazapentacosan-25-yl)hex-5-ynamide, ... | | Authors: | McGowan, S, Klemba, M, Greebaum, D.C. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2IA4
 
 | |
2GJ0
 
 | | Cycloviolacin O14 | | Descriptor: | Cycloviolacin O14 | | Authors: | Ireland, D.C, Colgrave, M.L, Craik, D.J. | | Deposit date: | 2006-03-30 | | Release date: | 2006-04-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | A novel suite of cyclotides from Viola odorata: sequence variation and the implications for structure, function and stability
Biochem.J., 400, 2006
|
|
2J17
 
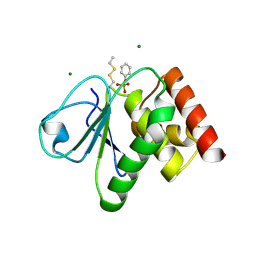 | | pTyr bound form of SDP-1 | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2IYB
 
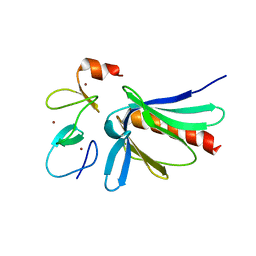 | |
2J16
 
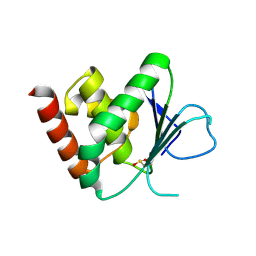 | | Apo & Sulphate bound forms of SDP-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2F5I
 
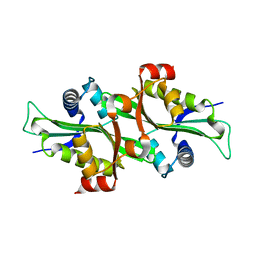 | |
2G3T
 
 | |
2F1G
 
 | | Cathepsin S in complex with non-covalent 2-(Benzoxazol-2-ylamino)-acetamide | | Descriptor: | Cathepsin S, GLYCEROL, N~2~-1,3-BENZOXAZOL-2-YL-3-CYCLOHEXYL-N-{2-[(4-METHOXYPHENYL)AMINO]ETHYL}-L-ALANINAMIDE | | Authors: | Spraggon, G, Hornsby, M, Lesley, S.A, Tully, D.C, Harris, J.L, Karenewsky, D.S, Kulathila, R, Clark, K. | | Deposit date: | 2005-11-14 | | Release date: | 2006-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of arylaminoethyl amides as noncovalent inhibitors of cathepsin S. Part 3: Heterocyclic P3.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3O2P
 
 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3O2U
 
 | | S. cerevisiae Ubc12 | | Descriptor: | GLYCEROL, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3OIQ
 
 | | Crystal structure of yeast telomere protein Cdc13 OB1 and the catalytic subunit of DNA polymerase alpha Pol1 | | Descriptor: | Cell division control protein 13, DNA polymerase alpha catalytic subunit A | | Authors: | Sun, J, Yang, Y, Wan, K, Mao, N, Yu, T.Y, Lin, Y.C, DeZwaan, D.C, Freeman, B.C, Lin, J.J, Lue, N.F, Lei, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha.
Cell Res., 21, 2011
|
|
3OY3
 
 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | Descriptor: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
3OIP
 
 | | Crystal structure of Yeast telomere protein Cdc13 OB1 | | Descriptor: | Cell division control protein 13 | | Authors: | Sun, J, Yang, Y, Wan, K, Mao, N, Yu, T.Y, Lin, Y.C, DeZwaan, D.C, Freeman, B.C, Lin, J.J, Lue, N.F, Lei, M. | | Deposit date: | 2010-08-19 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase alpha.
Cell Res., 21, 2011
|
|
3PSA
 
 | | Classification of a Haemophilus influenzae ABC transporter HI1470/71 through its cognate molybdate periplasmic binding protein MolA (MolA bound to tungstate) | | Descriptor: | TUNGSTATE(VI)ION, protein HI_1472 | | Authors: | Tirado-Lee, L, Lee, A, Rees, D.C, Pinkett, H.W. | | Deposit date: | 2010-12-01 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Classification of a Haemophilus influenzae ABC Transporter HI1470/71 through Its Cognate Molybdate Periplasmic Binding Protein, MolA.
Structure, 19, 2011
|
|
