6WX5
 
 | | Adducts formed after 3 weeks in the reaction of chlorido[chlorido(2,2'-((2-([2,2':6',2''-Terpyridin]-4'-yloxy)ethyl)azanediyl)bis(ethan-1-ol))platinum(II)] with HEWL | | Descriptor: | ACETATE ION, Lysozyme, PLATINUM (II) ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-05-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mustards-Derived Terpyridine-Platinum Complexes as Anticancer Agents: DNA Alkylation vs Coordination.
Inorg.Chem., 60, 2021
|
|
6WGO
 
 | | The interaction of dichlorido(3-(anthracen-9-ylmethyl)-1-methylimidazol-2-ylidene)(eta6-p-cymene)ruthenium(II) with HEWL after 1 week | | Descriptor: | ACETATE ION, Lysozyme, SODIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2020-04-06 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Anthracenyl Functionalization of Half-Sandwich Carbene Complexes: In Vitro Anticancer Activity and Reactions with Biomolecules.
Inorg.Chem., 60, 2021
|
|
2BF1
 
 | | Structure of an unliganded and fully-glycosylated SIV gp120 envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, B, Vogan, E.M, Gong, H, Skehel, J.J, Wiley, D.C, Harrison, S.C. | | Deposit date: | 2004-12-02 | | Release date: | 2005-02-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of an Unliganded Simian Immunodeficiency Virus Gp120 Core
Nature, 433, 2005
|
|
6XGY
 
 | | Crystal structure of E. coli MlaFB ABC transport subunits in the dimeric state | | Descriptor: | ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.
Elife, 9, 2020
|
|
6XKA
 
 | |
6XBD
 
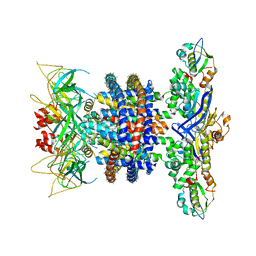 | | Cryo-EM structure of MlaFEDB in nanodiscs with phospholipid substrates | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, MSP1D1, Phospholipid ABC transporter permease protein MlaE, ... | | Authors: | Coudray, N, Isom, G.L, MacRae, M.R, Saiduddin, M, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure of bacterial phospholipid transporter MlaFEDB with substrate bound.
Elife, 9, 2020
|
|
6XGZ
 
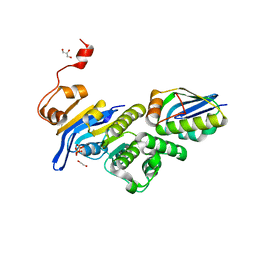 | | Crystal structure of E. coli MlaFB ABC transport subunits in the monomeric state | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | Authors: | Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.
Elife, 9, 2020
|
|
2ATC
 
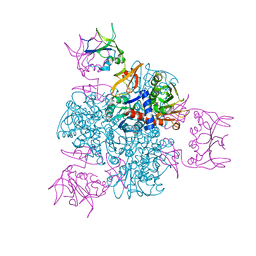 | | CRYSTAL AND MOLECULAR STRUCTURES OF NATIVE AND CTP-LIGANDED ASPARTATE CARBAMOYLTRANSFERASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE CARBAMOYLTRANSFERASE, CATALYTIC CHAIN, REGULATORY CHAIN, ... | | Authors: | Honzatko, R.B, Crawford, J.L, Monaco, H.L, Ladner, J.E, Edwards, B.F.P, Evans, D.R, Warren, S.G, Wiley, D.C, Ladner, R.C, Lipscomb, W.N. | | Deposit date: | 1982-03-24 | | Release date: | 1982-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli.
J.Mol.Biol., 160, 1982
|
|
6YXE
 
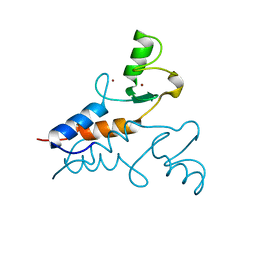 | | Structure of the Trim69 RING domain | | Descriptor: | E3 ubiquitin-protein ligase TRIM69, ZINC ION | | Authors: | Keown, J.R, Goldstone, D.C. | | Deposit date: | 2020-05-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The RING domain of TRIM69 promotes higher-order assembly.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7UDK
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | Descriptor: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDL
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | Descriptor: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | Descriptor: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDO
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
6BEJ
 
 | | Crystal structure of manganese superoxide dismutase from Xanthomonas citri | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Goto, L.S, Alexandrino, A.V, Pereira, C.M, Mendoca, D.C, Leonardo, D.A, Pereira, H.M, Garratt, R.C, Novo-Mansur, M.T.M. | | Deposit date: | 2017-10-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structural characterization of a pathogenicity-related superoxide dismutase codified by a probably essential gene in Xanthomonas citri subsp. citri.
PLoS ONE, 14, 2019
|
|
7UE2
 
 | |
7UPQ
 
 | |
7UJX
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 2.4 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-03-31 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7UPO
 
 | |
7UPP
 
 | |
7V0G
 
 | | Structure of cAMP-dependent protein kinase using a MD-MX procedure, produced using 1.63 Angstrom data | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wych, D.C, Aoto, P.C, Wall, M.E. | | Deposit date: | 2022-05-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Molecular-dynamics simulation methods for macromolecular crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6MFL
 
 | | Structure of siderophore binding protein BauB bound to a complex between two molecules of acinetobactin and ferric iron. | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Siderophore Binding Protein BauB, ... | | Authors: | Gulick, A.M, Bailey, D.C, Wencewicz, T.A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Siderophore Binding Protein BauB Bound to an Unusual 2:1 Complex Between Acinetobactin and Ferric Iron.
Biochemistry, 57, 2018
|
|
6MCW
 
 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, complex with the detergent Anapoe-X-114 | | Descriptor: | 23-[4-(2,4,4-trimethylpentan-2-yl)phenoxy]-3,6,9,12,15,18,21-heptaoxatricosan-1-ol, Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | Deposit date: | 2018-09-02 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
6MI0
 
 | | Crystal structure of the P450 domain of the CYP51-ferredoxin fusion protein from Methylococcus capsulatus, ligand-free state | | Descriptor: | Cytochrome P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hargrove, T, Wawrzak, Z, Lamb, D.C, Lepesheva, G.I. | | Deposit date: | 2018-09-18 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Concerning P450 evolution: Structural Analyses Support Bacterial Origin of Sterol 14 alpha-Demethylases.
Mol.Biol.Evol., 2020
|
|
6N4L
 
 | | Dithionite-reduced ADP-bound form of the nitrogenase Fe-protein from A. vinelandii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Wenke, B.B, Spatzal, T, Rees, D.C. | | Deposit date: | 2018-11-19 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Site-Specific Oxidation State Assignments of the Iron Atoms in the [4Fe:4S]2+/1+/0States of the Nitrogenase Fe-Protein.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
