3KYR
 
 | | Bace-1 in complex with a norstatine type inhibitor | | Descriptor: | 3-[[(2S)-2-[[[(2S)-2-[[(2S)-2-[[(2S)-2-azanyl-3-(1H-1,2,3,4-tetrazol-5-ylcarbonylamino)propanoyl]amino]-3-methyl-butanoyl]amino]-4-methyl-pentanoyl]amino]methyl]-2-hydroxy-4-phenyl-butanoyl]amino]benzoic acid, Beta-secretase 1 | | Authors: | Lindberg, J.D, Borkakoti, N, Derbyshire, D, Nystrom, S. | | Deposit date: | 2009-12-07 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of a-phenylnorstatine and a-benzylnorstatine as transition state isostere motifs in the search for new BACE-1 inhibiotrs
To be Published
|
|
3GG0
 
 | | Klebsiella pneumoniae BlrP1 pH 9.0 manganese/cy-diGMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FLAVIN MONONUCLEOTIDE, Klebsiella pneumoniae BlrP1, ... | | Authors: | Barends, T, Hartmann, E, Griese, J, Beitlich, T, Kirienko, N, Ryjenkov, D, Reinstein, J, Shoeman, R, Gomelsky, M, Schlichting, I. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase.
Nature, 459, 2009
|
|
1CIP
 
 | |
3GFY
 
 | | Klebsiella pneumoniae BlrP1 with FMN and cyclic diGMP, no metal ions | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FLAVIN MONONUCLEOTIDE, Klebsiella pneumoniae BlrP1 | | Authors: | Barends, T, Hartmann, E, Griese, J, Beitlich, T, Kirienko, N, Ryjenkov, D, Reinstein, J, Shoeman, R, Gomelsky, M, Schlichting, I. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase.
Nature, 459, 2009
|
|
2Q1S
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmF in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase | | Authors: | Harmer, N.J, King, J.D, Palmer, C.M, Maskell, D, Blundell, T.L. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2PZJ
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmF in complex with NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase | | Authors: | Harmer, N.J, King, J.D, Palmer, C.M, Maskell, D, Blundell, T.L. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2NPM
 
 | | crystal structure of Cryptosporidium parvum 14-3-3 protein in complex with peptide | | Descriptor: | 14-3-3 domain containing protein, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Dong, A, Lew, J, Wasney, G, Ren, H, Lin, L, Hassanali, A, Qiu, W, Zhao, Y, Doyle, D, Vedadi, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Brokx, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Characterization of 14-3-3 proteins from Cryptosporidium parvum.
Plos One, 6, 2011
|
|
3FM0
 
 | | Crystal structure of WD40 protein Ciao1 | | Descriptor: | Protein CIAO1, SULFATE ION | | Authors: | Dong, A, Ravichandran, M, Crombet, L, Cossar, D, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
2N8N
 
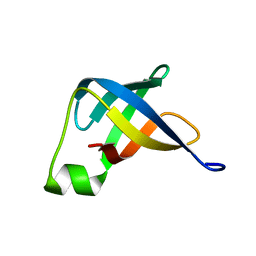 | | Solution structure of translation initiation factor | | Descriptor: | Translation initiation factor IF-1 | | Authors: | Kim, D, Lee, K, Lee, B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics study of translation initiation factor 1 from Staphylococcus aureus suggests its RNA binding mode
Biochim.Biophys.Acta, 1865, 2017
|
|
2N85
 
 | | NMR structure of OtTx1a - AMP in DPC micelles | | Descriptor: | Spiderine-1a | | Authors: | Nadezhdin, K, Romanovskaya, D, Sachkova, M, Vassilevski, A, Grishin, E, Kovalchuk, S, Arseniev, A. | | Deposit date: | 2015-10-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Modular toxin from the lynx spider Oxyopes takobius: Structure of spiderine domains in solution and membrane-mimicking environment.
Protein Sci., 26, 2017
|
|
2NLI
 
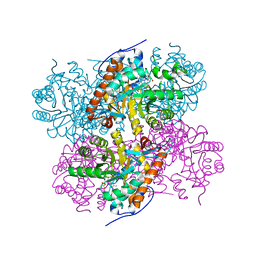 | | Crystal Structure of the complex between L-lactate oxidase and a substrate analogue at 1.59 angstrom resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, HYDROGEN PEROXIDE, LACTIC ACID, ... | | Authors: | Furuichi, M, Suzuki, N, Balasundaresan, D, Yoshida, Y, Minagawa, H, Watanabe, Y, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2006-10-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
3GZC
 
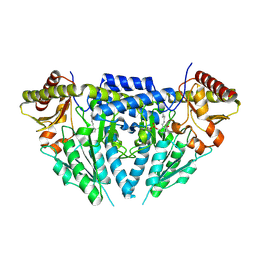 | | Structure of human selenocysteine lyase | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Selenocysteine lyase | | Authors: | Collins, R, Hogbom, M, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Karlberg, T, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
2Q9P
 
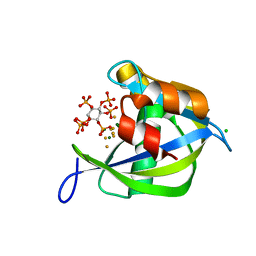 | | Human diphosphoinositol polyphosphate phosphohydrolase 1, Mg-F complex | | Descriptor: | CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, FLUORIDE ION, ... | | Authors: | Thorsell, A.G, Busam, R, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nordlund, P, Nyman, T, Ogg, D, Sagemark, J, Sundstrom, M, Van den Berg, S, Weigelt, J, Welin, M, Persson, C, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-13 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human diphosphoinositol phosphatase 1.
Proteins, 77, 2009
|
|
2Q1U
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmF in complex with NAD+ and UDP | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase, ... | | Authors: | Harmer, N.J, King, J.D, Palmer, C.M, Maskell, D, Blundell, T.L. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2PZM
 
 | | Crystal structure of the Bordetella bronchiseptica enzyme WbmG in complex with NAD and UDP | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative nucleotide sugar epimerase/ dehydratase, SULFATE ION, ... | | Authors: | Harmer, N.J, King, J.D, Palmer, C.M, Maskell, D, Blundell, T.L. | | Deposit date: | 2007-05-18 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Predicting protein function from structure--the roles of short-chain dehydrogenase/reductase enzymes in Bordetella O-antigen biosynthesis.
J.Mol.Biol., 374, 2007
|
|
2OT3
 
 | |
1DMA
 
 | | DOMAIN III OF PSEUDOMONAS AERUGINOSA EXOTOXIN COMPLEXED WITH NICOTINAMIDE AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, EXOTOXIN A, NICOTINAMIDE | | Authors: | Li, M, Dyda, F, Benhar, I, Pastan, I, Davies, D. | | Deposit date: | 1995-04-28 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Pseudomonas aeruginosa exotoxin domain III with nicotinamide and AMP: conformational differences with the intact exotoxin.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
2QFL
 
 | | Structure of SuhB: Inositol monophosphatase and extragenic suppressor from E. coli | | Descriptor: | ACETATE ION, ETHYL ACETATE, Inositol-1-monophosphatase | | Authors: | Wang, Y, Stieglitz, K.A, Bubunenko, M, Court, D, Stec, B, Roberts, M.F. | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the R184A mutant of the inositol monophosphatase encoded by suhB and implications for its functional interactions in Escherichia coli.
J.Biol.Chem., 282, 2007
|
|
1GWX
 
 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | 2-(4-{3-[1-[2-(2-CHLORO-6-FLUORO-PHENYL)-ETHYL]-3-(2,3-DICHLORO-PHENYL)-UREIDO]-PROPYL}-PHENOXY)-2-METHYL-PROPIONIC ACID, PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-17 | | Release date: | 2000-03-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
2MYH
 
 | | Omega-Tbo-IT1: selective inhibitor of insect calcium channels isolated from Tibellus oblongus spider venom | | Descriptor: | Omega-Tbo-IT1 toxin | | Authors: | Altukhov, D, Bozin, T, Bocharov, E, Kozlov, S, Mikov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-12-09 | | Method: | SOLUTION NMR | | Cite: | omega-Tbo-IT1-New Inhibitor of Insect Calcium Channels Isolated from Spider Venom.
Sci Rep, 5, 2015
|
|
2N1G
 
 | |
1H8D
 
 | | X-ray structure of the human alpha-thrombin complex with a tripeptide phosphonate inhibitor. | | Descriptor: | HIRUDIN I, N-[(benzyloxy)carbonyl]-beta-phenyl-D-phenylalanyl-N-{(1S,3E)-1-[dihydroxy(diphenoxy)-lambda~5~-phosphanyl]-4-methoxybut-3-en-1-yl}-L-prolinamide, THROMBIN | | Authors: | Skordalakes, E, Dodson, G.G, Green, D, Deadman, J. | | Deposit date: | 2001-02-01 | | Release date: | 2001-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of Human Alpha-Thrombin by a Phosphonate Tripeptide Proceeds Via a Metastable Pentacoordinated Phosphorus Intermediate
J.Mol.Biol., 311, 2001
|
|
2NBI
 
 | | Structure of the PSCD-region of the cell wall protein pleuralin-1 | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Rainer, D, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, H.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-12-21 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
1H0G
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argadin from Clonostachys | | Descriptor: | Argadin, CHITINASE B, GLYCEROL | | Authors: | Houston, D, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2QX3
 
 | |
