5V7N
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc04462 (SmGhrB) from Sinorhizobium meliloti in complex with NADP and 2-Keto-D-gluconic acid | | Descriptor: | 2-keto-D-gluconic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shabalin, I.G, Handing, K.B, Miks, C.D, Kutner, J, Matelska, D, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-03-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
5QON
 
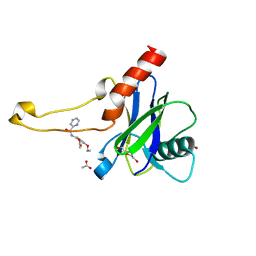 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000446a | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QOZ
 
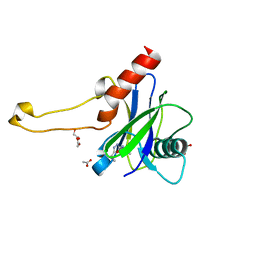 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with PB1787571279 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QOO
 
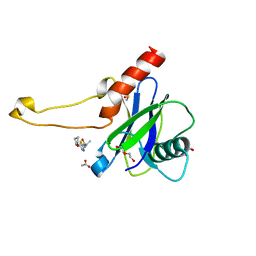 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000144a | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-N-{5-[(2S)-oxolan-2-yl]-1,3,4-thiadiazol-2(3H)-ylidene}propanamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QOU
 
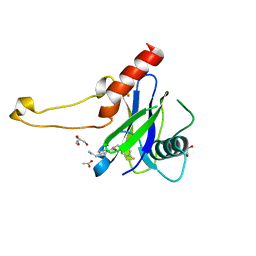 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z296300542 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QPA
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with FMOPL000449a | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-N-[(4-fluorophenyl)methyl]-1H-pyrazole-4-carboxamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QOQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z2895259680 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethyl-N-[(4-fluorophenyl)methyl]-1,2,4-oxadiazole-5-carboxamide, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QOY
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with YW-FY-378 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(2H-1,3-benzodioxole-5-carbonyl)-2,3,4,5-tetrahydro-1H-1,4-diazepin-1-yl]ethan-1-one, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QOR
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1203490773 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5OHN
 
 | |
5T4P
 
 | | Autoinhibited E. coli ATP synthase state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.77 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
5OCE
 
 | | THE MOLECULAR MECHANISM OF SUBSTRATE RECOGNITION AND CATALYSIS OF THE MEMBRANE ACYLTRANSFERASE PatA -- Complex of PatA with palmitate, mannose, and palmitoyl-6-mannose | | Descriptor: | PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase, [(2~{R},3~{S},4~{S},5~{S},6~{S})-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hexadecanoate, ... | | Authors: | Albesa-Jove, D, Tersa, M, Guerin, M.E. | | Deposit date: | 2017-06-30 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Molecular Mechanism of Substrate Recognition and Catalysis of the Membrane Acyltransferase PatA from Mycobacteria.
ACS Chem. Biol., 13, 2018
|
|
5OF8
 
 | | Cu nitrite reductase serial data at varying temperatures 190K 0.48MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5OEN
 
 | | Crystal Structure of STAT2 in complex with IRF9 | | Descriptor: | Interferon regulatory factor 9, Signal transducer and activator of transcription | | Authors: | Rengachari, S, Panne, D. | | Deposit date: | 2017-07-09 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.919 Å) | | Cite: | Structural basis of STAT2 recognition by IRF9 reveals molecular insights into ISGF3 function.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OFD
 
 | | Cu nitrite reductase serial data at varying temperatures 190K 0.48MGy | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5OEV
 
 | | The structure of a glutathione synthetase like-effector (GSS22) from Globodera pallida in apoform. | | Descriptor: | Glutathione synthetase-like effector 22 (Gpa-GSS22-apo) | | Authors: | Lilley, C.J, Maqbool, A, Wu, D, Yusup, H.B, Jones, L.M, Birch, P.R.J, Banfield, M.J, Urwin, P.E, Eves-van den Akker, S. | | Deposit date: | 2017-07-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Effector gene birth in plant parasitic nematodes: Neofunctionalization of a housekeeping glutathione synthetase gene.
PLoS Genet., 14, 2018
|
|
5OHL
 
 | | K6-specific affimer bound to K6 diUb | | Descriptor: | GLYCEROL, K6-specific affimer, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
5T0U
 
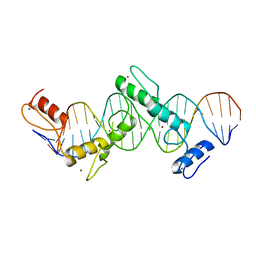 | | CTCF ZnF2-7 and DNA complex structure | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5T1J
 
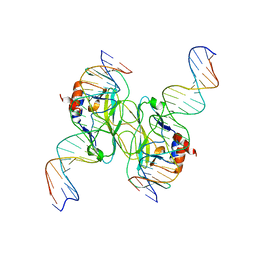 | | Crystal Structure of the Tbox DNA binding domain of the transcription factor T-bet | | Descriptor: | DNA, T-box transcription factor TBX21 | | Authors: | Liu, C.F, Brandt, G.S, Hoang, Q, Hwang, E.S, Naumova, N, Lazarevic, V, Dekker, J, Glimcher, L.H, Ringe, D, Petsko, G.A. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Crystal structure of the DNA binding domain of the transcription factor T-bet suggests simultaneous recognition of distant genome sites.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5OKO
 
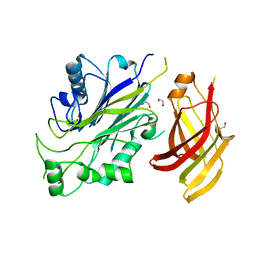 | | Crystal structure of human SHIP2 Phosphatase-C2 double mutant F593D/L597D | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2017-07-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for interdomain communication in SHIP2 providing high phosphatase activity.
Elife, 6, 2017
|
|
5OEU
 
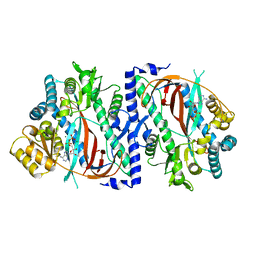 | | The structure of a glutathione synthetase like-effector (GSS22) from Globodera pallida in ADP-bound closed conformation. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutathione synthetase-like effector 22 (Gpa-GSS22-closed), MAGNESIUM ION | | Authors: | Lilley, C.J, Maqbool, A, Wu, D, Yusup, H.B, Jones, L.M, Birch, P.R.J, Banfield, M.J, Urwin, P.E, Eves-van den Akker, S. | | Deposit date: | 2017-07-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Effector gene birth in plant parasitic nematodes: Neofunctionalization of a housekeeping glutathione synthetase gene.
PLoS Genet., 14, 2018
|
|
5OCX
 
 | | Crystal structure of ACPA E4 in complex with CII-C-13-CIT | | Descriptor: | 1,2-ETHANEDIOL, CII-C-13-CIT, Fab fragment anti-citrullinated protein antibody E4 - light chain, ... | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5OEH
 
 | | Molecular tweezers modulate 14-3-3 protein-protein interactions. | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2017-07-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular tweezers modulate 14-3-3 protein-protein interactions.
Nat Chem, 5, 2013
|
|
5OHM
 
 | | K33-specific affimer bound to K33 diUb | | Descriptor: | K33-specific affimer, POLYETHYLENE GLYCOL (N=34), Polyubiquitin-C | | Authors: | Michel, M.A, Komander, D. | | Deposit date: | 2017-07-17 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Ubiquitin Linkage-Specific Affimers Reveal Insights into K6-Linked Ubiquitin Signaling.
Mol. Cell, 68, 2017
|
|
5OFH
 
 | | Cu nitrite reductase serial data at varying temperatures RT 0.15MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
