2NR4
 
 | | Crystal structure of FMN-bound protein MM1853 from Methanosarcina mazei, Pfam DUF447 | | Descriptor: | Conserved hypothetical protein, FLAVIN MONONUCLEOTIDE | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Lau, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-01 | | Release date: | 2006-11-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of conserved FMN bound hypothetical protein from Methanosarcina mazei
To be Published
|
|
1AB5
 
 | | STRUCTURE OF CHEY MUTANT F14N, V21T | | Descriptor: | CHEY | | Authors: | Wilcock, D, Pisabarro, M.T, Lopez-Hernandez, E, Serrano, L, Coll, M. | | Deposit date: | 1997-02-04 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure analysis of two CheY mutants: importance of the hydrogen-bond contribution to protein stability.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AJE
 
 | | CDC42 FROM HUMAN, NMR, 20 STRUCTURES | | Descriptor: | CDC42HS | | Authors: | Feltham, J.L, Dotsch, V, Raza, S, Manor, D, Cerione, R.A, Sutcliffe, M.J, Wagner, G, Oswald, R.E. | | Deposit date: | 1997-05-02 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Definition of the switch surface in the solution structure of Cdc42Hs.
Biochemistry, 36, 1997
|
|
2NUB
 
 | | Structure of Aquifex aeolicus Argonuate | | Descriptor: | Argonaute | | Authors: | Rashid, U.J, Paterok, D, Koglin, A, Gohlke, H, Piehler, J, Chen, J.C.-H. | | Deposit date: | 2006-11-09 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Aquifex aeolicus argonaute highlights conformational flexibility of the PAZ domain as a potential regulator of RNA-induced silencing complex function.
J.Biol.Chem., 282, 2007
|
|
2NXY
 
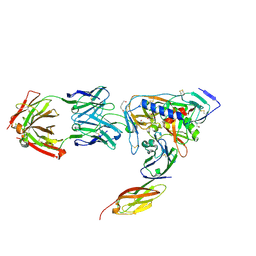 | | HIV-1 gp120 Envelope Glycoprotein(S334A) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
1ALV
 
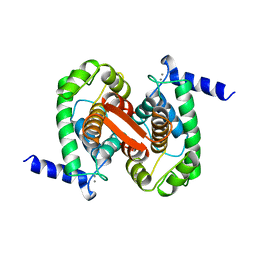 | | CALCIUM BOUND DOMAIN VI OF PORCINE CALPAIN | | Descriptor: | CALCIUM ION, CALPAIN | | Authors: | Narayana, S.V.L, Lin, G, Chattopadhyay, D, Maki, M. | | Deposit date: | 1997-06-03 | | Release date: | 1998-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of calcium bound domain VI of calpain at 1.9 A resolution and its role in enzyme assembly, regulation, and inhibitor binding.
Nat.Struct.Biol., 4, 1997
|
|
2NZJ
 
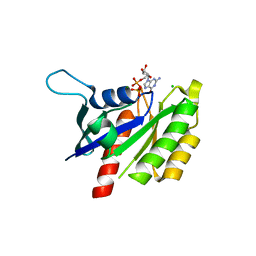 | | The crystal structure of REM1 in complex with GDP | | Descriptor: | CHLORIDE ION, GTP-binding protein REM 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Turnbull, A.P, Papagrigoriou, E, Ugochukwu, E, Elkins, J.M, Soundararajan, M, Yang, X, Gorrec, F, Umeano, C, Salah, E, Burgess, N, Johansson, C, Berridge, G, Gileadi, O, Bray, J, Marsden, B, Watts, S, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-23 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of REM1 in complex with GDP
To be Published
|
|
2O3X
 
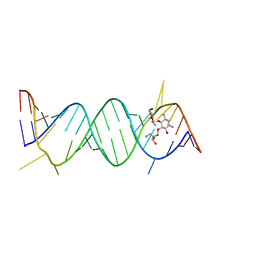 | | Crystal Structure of the Prokaryotic Ribosomal Decoding Site Complexed with Paromamine Derivative NB30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-DIAMINO-2-[(5-AMINO-5-DEOXY-BETA-D-RIBOFURANOSYL)OXY]-3-HYDROXYCYCLOHEXYL 2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSIDE, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | Deposit date: | 2006-12-02 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
2O45
 
 | | Structure of the 23S rRNA of the large ribosomal subunit from Deinococcus Radiodurans in complex with the macrolide RU-69874 | | Descriptor: | (3AS,4R,7R,8S,9S,10R,11R,13R,15R,15AR)-4-ETHYL-11-METHOXY-3A,7,9,11,13,15-HEXAMETHYL-2,6,14-TRIOXO-1-[4-(4-PYRIDIN-3-YL-1H-IMIDAZOL-1-YL)BUTYL]-10-{[3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSYL]OXY}TETRADECAHYDRO-2H-OXACYCLOTETRADECINO[4,3-D][1,3]OXAZOL-8-YL 2,6-DIDEOXY-3-C-METHYL-3-O-METHYL-ALPHA-L-RIBO-HEXOPYRANOSIDE, 23S rRNA | | Authors: | Baram, D, Pyetan, E, Auerbach-Nevo, T, Yonath, A. | | Deposit date: | 2006-12-03 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Chemical Parameters Influencing Fine Tuning in the Binding of Macrolide Antibiotics to the Ribosomal Tunnel
TO BE PUBLISHED
|
|
2Q7Z
 
 | | Solution Structure of the 30 SCR domains of human Complement Receptor 1 | | Descriptor: | Complement receptor type 1 | | Authors: | Furtado, P.B, Huang, C.Y, Ihyembe, D, Hammond, R.A, Marsh, H.C, Perkins, S.J. | | Deposit date: | 2007-06-08 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | The Partly Folded Back Solution Structure Arrangement of the 30 SCR Domains in Human Complement Receptor Type 1 (CR1) Permits Access to its C3b and C4b Ligands
J.Mol.Biol., 375, 2008
|
|
1A0D
 
 | | XYLOSE ISOMERASE FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | MANGANESE (II) ION, XYLOSE ISOMERASE | | Authors: | Gallay, O, Chopra, R, Conti, E, Brick, P, Blow, D. | | Deposit date: | 1997-11-28 | | Release date: | 1998-06-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Class II Xylose Isomerases from Two Thermophiles and a Hyperthermophile
To be Published
|
|
1A3P
 
 | | ROLE OF THE 6-20 DISULFIDE BRIDGE IN THE STRUCTURE AND ACTIVITY OF EPIDERMAL GROWTH FACTOR, NMR, 20 STRUCTURES | | Descriptor: | EPIDERMAL GROWTH FACTOR | | Authors: | Barnham, K, Torres, A, Alewood, D, Alewood, P, Domagala, T, Nice, E, Norton, R. | | Deposit date: | 1998-01-22 | | Release date: | 1998-07-29 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Role of the 6-20 disulfide bridge in the structure and activity of epidermal growth factor.
Protein Sci., 7, 1998
|
|
1COI
 
 | | DESIGNED TRIMERIC COILED COIL-VALD | | Descriptor: | COIL-VALD, SULFATE ION | | Authors: | Ogihara, N.L, Weiss, M.S, Degrado, W.F, Eisenberg, D. | | Deposit date: | 1996-08-10 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the designed trimeric coiled coil coil-VaLd: implications for engineering crystals and supramolecular assemblies.
Protein Sci., 6, 1997
|
|
1CGI
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE COMPLEXES BETWEEN BOVINE CHYMOTRYPSINOGEN*A AND TWO RECOMBINANT VARIANTS OF HUMAN PANCREATIC SECRETORY TRYPSIN INHIBITOR (KAZAL-TYPE) | | Descriptor: | ALPHA-CHYMOTRYPSINOGEN, PANCREATIC SECRETORY TRYPSIN INHIBITOR (KAZAL TYPE) VARIANT 3 | | Authors: | Hecht, H.J, Szardenings, M, Collins, J, Schomburg, D. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexes between bovine chymotrypsinogen A and two recombinant variants of human pancreatic secretory trypsin inhibitor (Kazal-type).
J.Mol.Biol., 220, 1991
|
|
1CGJ
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE COMPLEXES BETWEEN BOVINE CHYMOTRYPSINOGEN*A AND TWO RECOMBINANT VARIANTS OF HUMAN PANCREATIC SECRETORY TRYPSIN INHIBITOR (KAZAL-TYPE) | | Descriptor: | ALPHA-CHYMOTRYPSINOGEN, PANCREATIC SECRETORY TRYPSIN INHIBITOR (KAZAL TYPE) VARIANT 4 | | Authors: | Hecht, H.J, Szardenings, M, Collins, J, Schomburg, D. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of the complexes between bovine chymotrypsinogen A and two recombinant variants of human pancreatic secretory trypsin inhibitor (Kazal-type).
J.Mol.Biol., 220, 1991
|
|
1CV8
 
 | | STAPHOPAIN, CYSTEINE PROTEINASE FROM STAPHYLOCOCCUS AUREUS V8 | | Descriptor: | ACETATE ION, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, STAPHOPAIN | | Authors: | Hofmann, B, Schomburg, D, Hecht, H.-J. | | Deposit date: | 1998-05-08 | | Release date: | 1998-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of a Thiol Proteinase from Staphylococcus Aureus V-8 in the E-64 Inhibitor Complex
Acta Crystallogr.,Sect.A, 49, 1993
|
|
1CNO
 
 | | STRUCTURE OF PSEUDOMONAS NAUTICA CYTOCHROME C552, BY MAD METHOD | | Descriptor: | CYTOCHROME C552, GLYCEROL, HEME C | | Authors: | Brown, K, Nurizzo, D, Cambillau, C. | | Deposit date: | 1998-08-03 | | Release date: | 1999-07-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MAD structure of Pseudomonas nautica dimeric cytochrome c552 mimicks the c4 Dihemic cytochrome domain association.
J.Mol.Biol., 289, 1999
|
|
2Q91
 
 | | Structure of the Ca2+-Bound Activated Form of the S100A4 Metastasis Factor | | Descriptor: | CALCIUM ION, S100A4 Metastasis Factor | | Authors: | Malashkevich, V.N, Knight, D, Ramagopal, U.A, Almo, S.C, Bresnick, A.R. | | Deposit date: | 2007-06-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of Ca(2+)-Bound S100A4 and Its Interaction with Peptides Derived from Nonmuscle Myosin-IIA.
Biochemistry, 47, 2008
|
|
2Q9E
 
 | | Structure of spin-labeled T4 lysozyme mutant S44R1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (2.1 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
1D9P
 
 | |
2Q6U
 
 | | SeMet-substituted form of NikD | | Descriptor: | BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NikD protein | | Authors: | Carrell, C.J, Bruckner, R.C, Venci, D, Zhao, G, Jorns, M.S, Mathews, F.S. | | Deposit date: | 2007-06-05 | | Release date: | 2007-07-31 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NikD, an Unusual Amino Acid Oxidase Essential for Nikkomycin Biosynthesis: Structures of Closed and Open Forms at 1.15 and 1.90 A Resolution
Structure, 15, 2007
|
|
2Q7Q
 
 | |
1DKM
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DN5
 
 | | SOLVATION OF THE LEFT-HANDED HEXAMER D(5BRC-G-5BRC-G-5BRC-G) IN CRYSTALS GROWN AT TWO TEMPERATURES | | Descriptor: | DNA (5'-D(*(CBR)P*GP*(CBR)P*GP*(CBR)P*G)-3') | | Authors: | Chevrier, B, Dock, A.C, Hartmann, B, Leng, M, Moras, D, Thuong, M.T, Westhof, E. | | Deposit date: | 1986-12-01 | | Release date: | 1987-04-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Solvation of the left-handed hexamer d(5BrC-G-5BrC-G-5 BrC-G) in crystals grown at two temperatures.
J.Mol.Biol., 188, 1986
|
|
1DB2
 
 | | CRYSTAL STRUCTURE OF NATIVE PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Nar, H, Bauer, M, Stassen, J.M, Lang, D, Gils, A, Declerck, P. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation.
J.Mol.Biol., 297, 2000
|
|
