7ZSU
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, Steroid C26-monooxygenase, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
2WEZ
 
 | | Human BACE-1 in complex with 1-ethyl-N-((1S,2R)-2-hydroxy-3-(((3-(methyloxy)phenyl)methyl)amino)-1-(phenylmethyl)propyl)-4-(2-oxo-1- pyrrolidinyl)-1H-indole-6-carboxamide | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-BENZYL-2-HYDROXY-3-[(3-METHOXYBENZYL)AMINO]PROPYL}-1-ETHYL-4-(2-OXOPYRROLIDIN-1-YL)-1H-INDOLE-6-CARBOXAMIDE | | Authors: | Charrier, N, Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, Hawkins, J, Howes, C, Hubbard, J, Hussain, I, Maile, G, Matico, R, Mosley, J, Naylor, A, O'Brien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-19 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Second Generation of Bace-1 Inhibitors. Part 1: The Need for Improved Pharmacokinetics.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7ZLZ
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, Steroid C26-monooxygenase, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
2WQZ
 
 | | Crystal structure of synaptic protein neuroligin-4 in complex with neurexin-beta 1: alternative refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEUREXIN-1-BETA, ... | | Authors: | Fabrichny, I.P, Leone, P, Sulzenbacher, G, Comoletti, D, Miller, M.T, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Analysis of the Synaptic Protein Neuroligin and its Beta-Neurexin Complex: Determinants for Folding and Cell Adhesion.
Neuron, 56, 2007
|
|
7DWV
 
 | | Cryo-EM structure of amyloid fibril formed by familial prion disease-related mutation E196K | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Li, X.N, Dang, H.B, Ma, Y.Y, Wang, Q, Wang, C, Sun, Y.P, Chen, J, Li, D, Zhang, D.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2021-01-18 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Genetic prion disease-related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
2IDF
 
 | | P. aeruginosa azurin N42C/M64E double mutant, BMME-linked dimer | | Descriptor: | 1-[PYRROL-1-YL-2,5-DIONE-METHOXYMETHYL]-PYRROLE-2,5-DIONE, Azurin, COPPER (II) ION, ... | | Authors: | Einsle, O, de Jongh, T.E, Hoffmann, M, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-09-15 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electron transfer in a crosslinked protein dimer mediated by a hydrogen-bonded network across the dimer interface
To be Published
|
|
7ZW6
 
 | | Oligomeric structure of SynDLP | | Descriptor: | Slr0869 protein | | Authors: | Gewehr, L, Junglas, B, Jilly, R, Franz, J, Wenyu, E.Z, Weidner, T, Bonn, M, Sachse, C, Schneider, D. | | Deposit date: | 2022-05-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | SynDLP is a dynamin-like protein of Synechocystis sp. PCC 6803 with eukaryotic features.
Nat Commun, 14, 2023
|
|
2WHT
 
 | |
2X06
 
 | | SULFOLACTATE DEHYDROGENASE FROM METHANOCALDOCOCCUS JANNASCHII | | Descriptor: | L-SULFOLACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2009-12-07 | | Release date: | 2009-12-15 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Methanoarchaeal Sulfolactate Dehydrogenase: Prototype of a New Family of Nadh-Dependent Enzymes.
Embo J., 23, 2004
|
|
2X8X
 
 | |
1OPJ
 
 | | Structural basis for the auto-inhibition of c-Abl tyrosine kinase | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Young, M.A, Scheffzek, K, Veach, D, Bornmann, W, Clarkson, B, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the autoinhibition of c-Abl tyrosine kinase
Cell(Cambridge,Mass.), 112, 2003
|
|
2X93
 
 | | Crystal structure of AnCE-trandolaprilat complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ANGIOTENSIN CONVERTING ENZYME, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
2WX1
 
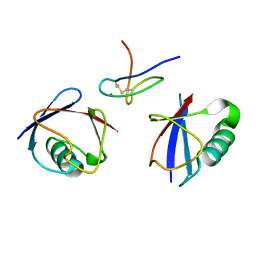 | | TAB2 NZF DOMAIN IN COMPLEX WITH Lys63-linked tri-ubiquitin, P212121 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 2, UBIQUITIN, ZINC ION | | Authors: | Kulathu, Y, Akutsu, M, Bremm, A, Hofmann, K, Komander, D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-Sided Ubiquitin Binding Explains Specificity of the Tab2 Nzf Domain
Nat.Struct.Mol.Biol., 16, 2009
|
|
1OIG
 
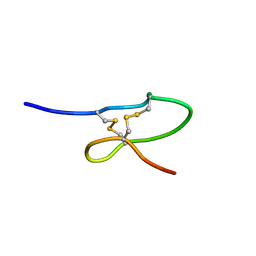 | | The solution structure of the DPY module from the Dumpy protein | | Descriptor: | Dumpy, isoform Y | | Authors: | Wilkin, M.B, Becker, M.N, Mulvey, D, Phan, I, Chao, A, Cooper, K, Chung, H.J, Campbell, I.D, Baron, M, MacIntyre, R. | | Deposit date: | 2003-06-18 | | Release date: | 2003-06-26 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Drosophila Dumpy is a Gigantic Extracellular Protein Required to Maintain Tension at Epidermal-Cuticle Attachment Sites
Curr.Biol., 10, 2000
|
|
1OHQ
 
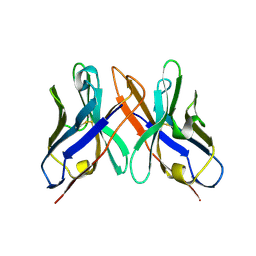 | | Crystal structure of HEL4, a soluble human VH antibody domain resistant to aggregation | | Descriptor: | IMMUNOGLOBULIN | | Authors: | Jespers, L, Schon, O, James, L.C, Veprintsev, D, Winter, G. | | Deposit date: | 2003-05-30 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hel4, a Soluble, Refoldable Human V(H) Single Domain with a Germ-Line Scaffold
J.Mol.Biol., 337, 2004
|
|
7ZJS
 
 | |
7NLC
 
 | | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide | | Descriptor: | AMMONIUM ION, CHLORIDE ION, Protein ORF3, ... | | Authors: | Moschidi, D, Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Crystallographic structure of human Tsg101 UEV domain in complex with a HEV ORF3 peptide
To Be Published
|
|
1OM2
 
 | | SOLUTION NMR STRUCTURE OF THE MITOCHONDRIAL PROTEIN IMPORT RECEPTOR TOM20 FROM RAT IN A COMPLEX WITH A PRESEQUENCE PEPTIDE DERIVED FROM RAT ALDEHYDE DEHYDROGENASE (ALDH) | | Descriptor: | PROTEIN (MITOCHONDRIAL ALDEHYDE DEHYDROGENASE), PROTEIN (MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20) | | Authors: | Abe, Y, Shodai, T, Muto, T, Mihara, K, Torii, H, Nishikawa, S, Endo, T, Kohda, D. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20.
Cell(Cambridge,Mass.), 100, 2000
|
|
7ND2
 
 | | Cryo-EM structure of the human FERRY complex | | Descriptor: | Glutamine amidotransferase-like class 1 domain-containing protein 1, Protein phosphatase 1 regulatory subunit 21, Quinone oxidoreductase-like protein 1 | | Authors: | Quentin, D, Klink, B.U, Raunser, S. | | Deposit date: | 2021-01-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mRNA binding by the human FERRY Rab5 effector complex.
Mol.Cell, 83, 2023
|
|
2X31
 
 | | Modelling of the complex between subunits BchI and BchD of magnesium chelatase based on single-particle cryo-EM reconstruction at 7.5 ang | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT, MAGNESIUM-CHELATASE 60 KDA SUBUNIT | | Authors: | Lunqvist, J, Elmlund, H, Peterson Wulff, R, Berglund, L, Elmlund, D, Emanuelsson, C, Hebert, H, Willows, R.D, Hansson, M, Lindahl, M, Al-Karadaghi, S. | | Deposit date: | 2010-01-19 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | ATP-Induced Conformational Dynamics in the Aaa+ Motor Unit of Magnesium Chelatase.
Structure, 18, 2010
|
|
1ONA
 
 | | CO-CRYSTALS OF CONCANAVALIN A WITH METHYL-3,6-DI-O-(ALPHA-D-MANNOPYRANOSYL)-ALPHA-D-MANNOPYRANOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Bouckaert, J, Maes, D, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1996-07-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structure of the complex between concanavalin A and methyl-3,6-di-O-(alpha-D-mannopyranosyl)-alpha-D-mannopyranoside reveals two binding modes.
J.Biol.Chem., 271, 1996
|
|
4HPX
 
 | | Crystal structure of Tryptophan Synthase at 1.65 A resolution in complex with alpha aminoacrylate E(A-A) and benzimidazole in the beta site and the F9 inhibitor in the alpha site | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
1OOE
 
 | | Structural Genomics of Caenorhabditis elegans : Dihydropteridine reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydropteridine reductase | | Authors: | Symersky, J, Li, S, Nagy, L, Qiu, S, Lin, G, Tsao, J, Luo, D, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of dihydropteridine reductase.
Proteins, 53, 2003
|
|
7ZPN
 
 | | Crystal Structure of IscR from Dinoroseobacter shibae | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator, SULFATE ION | | Authors: | Lukat, P, Ploetzky, L, Blankenfeldt, W, Jahn, D, Haertig, E. | | Deposit date: | 2022-04-28 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Dinoroseobacter shibae IscR homolog acts as a repressor for iron acquisition genes
To Be Published
|
|
1OP8
 
 | | Crystal Structure of Human Granzyme A | | Descriptor: | Granzyme A, SULFATE ION | | Authors: | Hink-Schauer, C, Estebanez-Perpina, E, Bode, W, Jenne, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the apoptosis-inducing human granzyme A dimer
NAT.STRUCT.BIOL., 10, 2003
|
|
