5E1S
 
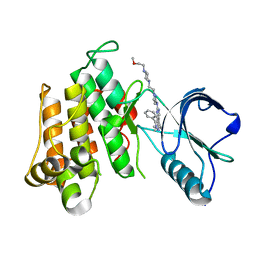 | | The Crystal structure of INSR Tyrosine Kinase in complex with the Inhibitor BI 885578 | | Descriptor: | (5R)-N-(1-{2-[4-(2-methoxyethyl)piperazin-1-yl]ethyl}-1H-pyrazol-3-yl)-5,8-dimethyl-9-phenyl-6,8-dihydro-5H-pyrazolo[3,4-h]quinazolin-2-amine, Insulin receptor | | Authors: | Kessler, D, Zahn, S, Sanderson, M, Wolkerstorfer, B. | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | BI 885578, a Novel IGF1R/INSR Tyrosine Kinase Inhibitor with Pharmacokinetic Properties That Dissociate Antitumor Efficacy and Perturbation of Glucose Homeostasis.
Mol.Cancer Ther., 14, 2015
|
|
5E1Z
 
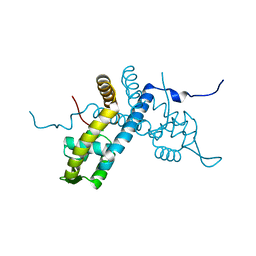 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 2,4-dichlorophenol bound form | | Descriptor: | 2,4-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
to be published
|
|
1TG8
 
 | | The structure of Dengue virus E glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, envelope glycoprotein | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2004-05-28 | | Release date: | 2004-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
1THD
 
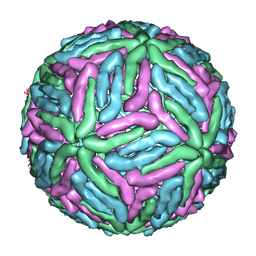 | | COMPLEX ORGANIZATION OF DENGUE VIRUS E PROTEIN AS REVEALED BY 9.5 ANGSTROM CRYO-EM RECONSTRUCTION | | Descriptor: | Major envelope protein E | | Authors: | Zhang, Y, Zhang, W, Ogata, S, Clements, D, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2004-06-01 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Conformational changes of the flavivirus e glycoprotein
Structure, 12, 2004
|
|
5HFS
 
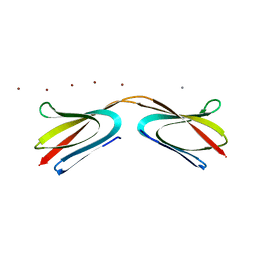 | | CRYSTAL STRUCTURE OF C-TERMINAL DOMAIN OF CARGO PROTEINS OF TYPE IX SECRETION SYSTEM | | Descriptor: | CALCIUM ION, Gingipain R2, ZINC ION | | Authors: | Golik, P, Szmigielski, B, Ksiazek, M, Nowakowska, Z, Mizgalska, D, Nowak, M, Dubin, G, Potempa, J. | | Deposit date: | 2016-01-07 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain.
Sci Rep, 6, 2016
|
|
1SU3
 
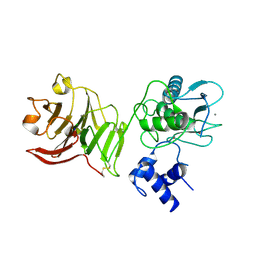 | | X-ray structure of human proMMP-1: New insights into collagenase action | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jozic, D, Bourenkov, G, Lim, N.H, Nagase, H, Bode, W, Maskos, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-26 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of human proMMP-1: new insights into procollagenase activation and collagen binding.
J.Biol.Chem., 280, 2005
|
|
1TD3
 
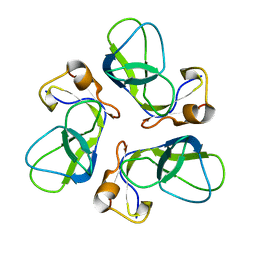 | | Crystal structure of VSHP_BPP21 in space group C2 | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP.
J.Mol.Biol., 344, 2004
|
|
1SWW
 
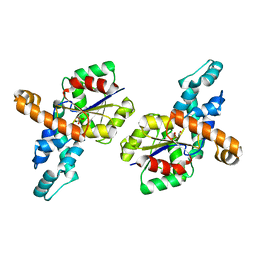 | | Crystal structure of the phosphonoacetaldehyde hydrolase D12A mutant complexed with magnesium and substrate phosphonoacetaldehyde | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETALDEHYDE, phosphonoacetaldehyde hydrolase | | Authors: | Zhang, G, Morais, M.C, Dai, J, Zhang, W, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of metal ion binding in phosphonoacetaldehyde hydrolase identifies sequence markers for metal-activated enzymes of the HAD enzyme superfamily
Biochemistry, 43, 2004
|
|
3KVR
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | 2,5-anhydro-4-deoxy-D-erythro-pent-4-enitol, 5-FLUOROURACIL, SULFATE ION, ... | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-30 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
5HTC
 
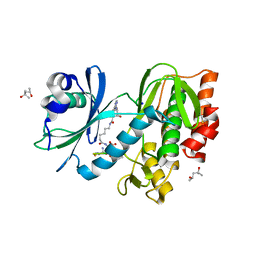 | | Crystal structure of haspin (GSG2) in complex with bisubstrate inhibitor ARC-3372 | | Descriptor: | (2R)-2-{[6-({[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]carbonyl}amino)hexanoyl]amino}butanedioic acid (non-preferred name), (4S)-2-METHYL-2,4-PENTANEDIOL, ARC-3372 INHIBITOR, ... | | Authors: | Chaikuad, A, Heroven, C, Lavogina, D, Kestav, K, Uri, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-01-26 | | Release date: | 2016-03-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structures of the protein kinase haspin with bisubstrate inhibitors.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1V6C
 
 | | Crystal Structure of Psychrophilic Subtilisin-like Protease Apa1 from Antarctic Psychrotroph Pseudoalteromonas sp. AS-11 | | Descriptor: | CALCIUM ION, SULFATE ION, alkaline serine protease, ... | | Authors: | Dong, D, Ihara, T, Motoshima, H, Watanabe, K. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Psychrophilic Subtilisin-like Protease Apa1 from Antarctic Psychrotroph Pseudoalteromonas sp. AS-11
To be Published
|
|
5EA7
 
 | | Crystal Structure of Inhibitor BMS-433771 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1-cyclopropyl-3-[[1-(4-oxidanylbutyl)benzimidazol-2-yl]methyl]imidazo[4,5-c]pyridin-2-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
5HX1
 
 | |
1BD0
 
 | | ALANINE RACEMASE COMPLEXED WITH ALANINE PHOSPHONATE | | Descriptor: | ALANINE RACEMASE, {1-[(3-HYDROXY-METHYL-5-PHOSPHONOOXY-METHYL-PYRIDIN-4-YLMETHYL)-AMINO]-ETHYL}-PHOSPHONIC ACID | | Authors: | Stamper, G.F, Morollo, A.A, Ringe, D. | | Deposit date: | 1998-05-12 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine.
Biochemistry, 37, 1998
|
|
5HMQ
 
 | |
5E0B
 
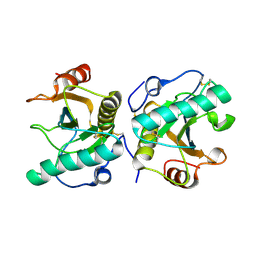 | | Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, N-acetyl-beta-muramic acid, ... | | Authors: | Sharma, P, Yamini, S, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-09-28 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the complex of Peptidoglycan recognition protein PGRP-S with N-Acetyl Muramic acid at 2.6 A resolution
To Be Published
|
|
1UU8
 
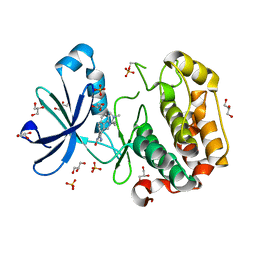 | | Structure of human PDK1 kinase domain in complex with BIM-1 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Schuttelkopf, A.W, Deak, M, Prakash, K.R, Bain, J, Elliot, M, Garrido-Franco, M, Kozikowski, A.P, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-12-16 | | Release date: | 2004-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interactions of Ly333531 and Other Bisindolyl Maleimide Inhibitors with Pdk1
Structure, 12, 2004
|
|
3L2X
 
 | |
5DP2
 
 | |
5E3O
 
 | |
3NM3
 
 | | The Crystal Structure of Candida glabrata THI6, a Bifunctional Enzyme involved in Thiamin Biosyhthesis of Eukaryotes | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, THIAMIN PHOSPHATE, ... | | Authors: | Paul, D, Chatterjee, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2010-06-21 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Domain Organization in Candida glabrata THI6, a Bifunctional Enzyme Required for Thiamin Biosynthesis in Eukaryotes .
Biochemistry, 49, 2010
|
|
5DS9
 
 | |
5E5X
 
 | |
5HA7
 
 | | Human Aldose Reductase in Complex with NADP+ and WY14643 in Space Group P212121 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sawaya, M.R, Cascio, D, Balendiran, G.K. | | Deposit date: | 2015-12-30 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Characterization of WY 14,643 and its Complex with Aldose Reductase.
Sci Rep, 6, 2016
|
|
5E7D
 
 | | Crystal Structure of the fifth bromodomain of human PB1 in complex with a hydroxyphenyl ligand | | Descriptor: | (2E)-3-(dimethylamino)-1-(2-hydroxyphenyl)prop-2-en-1-one, 1,2-ETHANEDIOL, Protein polybromo-1 | | Authors: | Filippakopoulos, P, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Owen, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-12 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of the fifth bromodomain of human PB1 in complex with a hydroxyphenyl ligand
To Be Published
|
|
