5WE8
 
 | |
2RMK
 
 | | Rac1/PRK1 Complex | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1, ... | | Authors: | Modha, R, Campbell, L.J, Nietlispach, D, Buhecha, H.R, Owen, D, Mott, H.R. | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Rac1 Polybasic Region Is Required for Interaction with Its Effector PRK1
J.Biol.Chem., 283, 2008
|
|
4PV2
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with K+ and Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, NITRATE ION, ... | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Clavel, D, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2RLP
 
 | | NMR structure of CCP modules 1-2 of complement factor H | | Descriptor: | Complement factor H | | Authors: | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | Deposit date: | 2007-07-28 | | Release date: | 2008-02-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
3CN7
 
 | |
3CSC
 
 | |
3VK3
 
 | | Crystal Structure of L-Methionine gamma-Lyase from Pseudomonas putida C116H Mutant Complexed with L-methionine | | Descriptor: | METHIONINE, Methionine gamma-lyase | | Authors: | Fukumoto, M, Kudou, D, Murano, S, Shiba, T, Sato, D, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2011-11-07 | | Release date: | 2012-09-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of amino acid residues in the active site of L-methionine gamma-lyase from Pseudomonas putida.
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3VK4
 
 | | Crystal Structure of L-Methionine gamma-Lyase from Pseudomonas putida C116H Mutant complexed with L-homocysteine | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Methionine gamma-lyase | | Authors: | Fukumoto, M, Kudou, D, Murano, S, Shiba, T, Sato, D, Tamura, T, Harada, S, Inagaki, K. | | Deposit date: | 2011-11-07 | | Release date: | 2012-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The role of amino acid residues in the active site of L-methionine gamma-lyase from Pseudomonas putida.
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3LIP
 
 | |
3LQS
 
 | | Complex Structure of D-Amino Acid Aminotransferase and 4-amino-4,5-dihydro-thiophenecarboxylic acid (ADTA) | | Descriptor: | 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, ACETIC ACID, D-alanine aminotransferase | | Authors: | Lepore, B.W, Liu, D, Peng, Y, Fu, M, Yasuda, C, Manning, J.M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chiral discrimination among aminotransferases: inactivation by 4-amino-4,5-dihydrothiophenecarboxylic acid.
Biochemistry, 49, 2010
|
|
5HO7
 
 | | DISCOVERY OF NOVEL 7-AZAINDOLES AS PDK1 INHIBITORS | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, 5-amino-3-(methylsulfanyl)-1H-pyrazole-1,4-dicarboxamide, SULFATE ION | | Authors: | Wucherer-Plietker, M, Esdar, C, Knoechel, T, Hillertz, P, Heinrich, T, Buchstaller, H.P, Greiner, H, Dorsch, D, Calderini, M, Bruge, D, Mueller, T.J.J, Graedler, U. | | Deposit date: | 2016-01-19 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of novel 7-azaindoles as PDK1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7N8I
 
 | |
7N8H
 
 | |
3MML
 
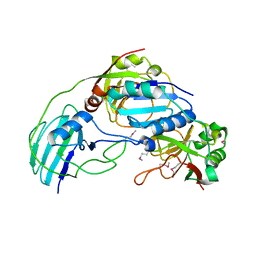 | | Allophanate Hydrolase Complex from Mycobacterium smegmatis, Msmeg0435-Msmeg0436 | | Descriptor: | Allophanate hydrolase subunit 1, Allophanate hydrolase subunit 2, CHLORIDE ION | | Authors: | Kaufmann, M, Chernishof, I, Shin, A, Germano, D, Sawaya, M.R, Waldo, G.S, Arbing, M.A, Perry, J, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-04-20 | | Release date: | 2010-04-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Allphanate Hydrolase Complex from M. smegmatis, Msmeg0435-Msmeg0436
To be Published
|
|
5IM5
 
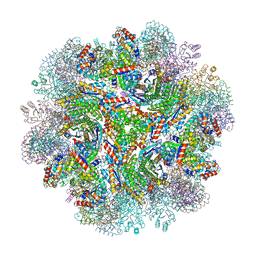 | | Crystal structure of designed two-component self-assembling icosahedral cage I53-40 | | Descriptor: | Designed Keto-hydroxyglutarate-aldolase/keto-deoxy-phosphogluconate aldolase, Designed Riboflavin synthase | | Authors: | Liu, Y.A, Cascio, D, Sawaya, M.R, Bale, J.B, Collazo, M.J, Thomas, C, Sheffler, W, King, N.P, Baker, D, Yeates, T.O. | | Deposit date: | 2016-03-05 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.699 Å) | | Cite: | Accurate design of megadalton-scale two-component icosahedral protein complexes.
Science, 353, 2016
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V2C
 
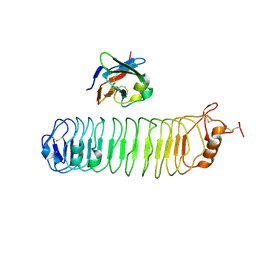 | | mouse FLRT2 LRR domain in complex with rat Unc5D Ig1 domain | | Descriptor: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 2, PROTEIN UNC5D | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
4UUR
 
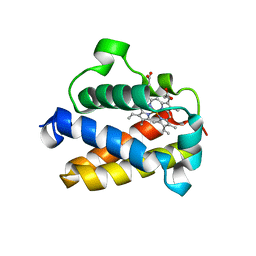 | | Cold-adapted truncated hemoglobin from the Antarctic marine bacterium Pseudoalteromonas haloplanktis TAC125 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE HEMOGLOBIN-LIKE OXYGEN-BINDING PROTEIN | | Authors: | Pesce, A, Giordano, D, Riccio, A, Nardini, M, Caldelli, E, Howes, B, Bustamante, J.P, Boechi, L, Estrin, D, di Prisco, G, Smulevich, G, Verde, C, Bolognesi, M. | | Deposit date: | 2014-07-31 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Flexibility of the Heme Cavity in the Cold-Adapted Truncated Hemoglobin from the Antarctic Marine Bacterium Pseudoalteromonas Haloplanktis Tac125.
FEBS J., 282, 2015
|
|
4UXW
 
 | | Structure of delta4-DgkA-apo in 9.9 MAG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4S)-2-METHYL-2,4-PENTANEDIOL, DIACYLGLYCEROL KINASE, ... | | Authors: | Li, D, Pye, V.E, Aragao, D, Caffrey, M. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase.
Nat.Commun., 6, 2015
|
|
4UUQ
 
 | | Crystal structure of human mono-glyceride lipase in complex with SAR127303 | | Descriptor: | 4-({[(4-chlorophenyl)sulfonyl]amino}methyl)piperidine-1-carboxylic acid, MONOGLYCERIDE LIPASE | | Authors: | Griebel, G, Pichat, P, Beeske, S, Leroy, T, Redon, N, Francon, D, Bert, L, Even, L, Lopez-Grancha, M, Tolstykh, T, Sun, F, Yu, Q, Brittain, S, Arlt, H, He, T, Zhang, B, Wiederschain, D, Bertrand, T, Houtman, J, Rak, A, Vallee, F, Michot, N, Auge, F, Menet, V, Bergis, O.E, George, P, Avenet, P, Mikol, V, Didier, M, Escoubet, J. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Selective Blockade of the Hydrolysis of the Endocannabinoid 2-Arachidonoylglycerol Impairs Learning and Memory Performance While Producing Antinociceptive Activity in Rodents.
Sci.Rep., 5, 2015
|
|
4V2E
 
 | | FLRT3 LRR domain | | Descriptor: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 3 | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development.
Neuron, 84, 2014
|
|
4WFS
 
 | | Crystal Structure of tRNA-dihydrouridine(20) synthase catalytic domain | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
1ELG
 
 | | NATURE OF THE INACTIVATION OF ELASTASE BY N-PEPTIDYL-O-AROYL HYDROXYLAMINE AS A FUNCTION OF PH | | Descriptor: | (TERT-BUTYLOXYCARBONYL)-ALANYL-ALANYL-AMINE, CALCIUM ION, PORCINE PANCREATIC ELASTASE | | Authors: | Ding, X, Rasmussen, B, Demuth, H.-U, Ringe, D, Steinmetz, A.C.U. | | Deposit date: | 1995-03-13 | | Release date: | 1995-07-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nature of the inactivation of elastase by N-peptidyl-O-aroyl hydroxylamine as a function of pH.
Biochemistry, 34, 1995
|
|
1EVL
 
 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH A THREONYL ADENYLATE ANALOG | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
7ZBE
 
 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SwissFEL) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
