2XWS
 
 | | ANAEROBIC COBALT CHELATASE (CbiX) FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | SIROHYDROCHLORIN COBALTOCHELATASE | | Authors: | Romao, C.V, Ladakis, D, Lobo, S.A.L, Carrondo, M.A, Brindley, A.A, Deery, E, Matias, P.M, Pickersgill, R.W, Saraiva, L.M, Warren, M.J. | | Deposit date: | 2010-11-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3GN4
 
 | | Myosin lever arm | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Mukherjea, M, Llinas, P, Kim, H, Travaglia, M, Safer, D, Zong, A.B, Menetrey, J, Franzini-Armstrong, C, Selvin, P.R, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2009-03-16 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach
Mol.Cell, 35, 2009
|
|
3GTO
 
 | | Backtracked RNA polymerase II complex with 15mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
2XWQ
 
 | | Anaerobic cobalt chelatase from Archeaoglobus fulgidus (CbiX) in complex with metalated sirohydrochlorin product | | Descriptor: | COBALT SIROHYDROCHLORIN, SIROHYDROCHLORIN COBALTOCHELATASE | | Authors: | Ladakis, D, Brindley, A.A, Deery, E, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2010-11-04 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Evolution in a Family of Chelatases Facilitated by the Introduction of Active Site Asymmetry and Protein Oligomerization.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XFQ
 
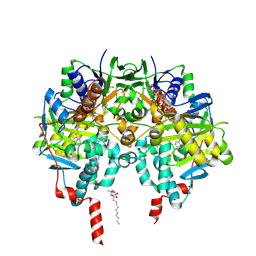 | | Rasagiline-inhibited human monoamine oxidase B in complex with 2-(2- benzofuranyl)-2-imidazoline | | Descriptor: | (1R)-N-(prop-2-en-1-yl)-2,3-dihydro-1H-inden-1-amine, 2-(2-BENZOFURANYL)-2-IMIDAZOLINE, Amine oxidase [flavin-containing] B, ... | | Authors: | Bonivento, D, Milczek, E.M, McDonald, G.R, Binda, C, Holt, A, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potentiation of ligand binding through cooperative effects in monoamine oxidase B.
J. Biol. Chem., 285, 2010
|
|
3GSO
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV peptide | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503 (NLVPMVATV), HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Saulquin, X, Reiser, J.-B, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
2XBR
 
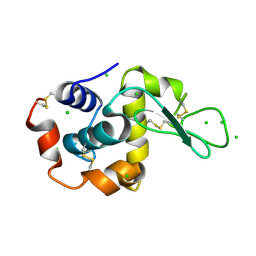 | | Raman crystallography of Hen White Egg Lysozyme - Low X-ray dose (0.2 MGy) | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Carpentier, P, Royant, A, Weik, M, Bourgeois, D. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Raman Assisted Crystallography Reveals a Mechanism of X-Ray Induced Reversible Disulfide Radical Formation
Structure, 18, 2010
|
|
3GTQ
 
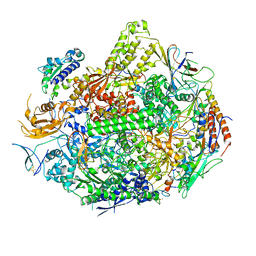 | | Backtracked RNA polymerase II complex induced by damage | | Descriptor: | DNA (5'-D(*CP*TP*AP*CP*CP*CP*AP*TP*AP*AP*CP*CP*AP*CP*AP*GP*GP*CP*TP*CP*CP*TP*CP*TP*CP*CP*AP*TP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
2XP8
 
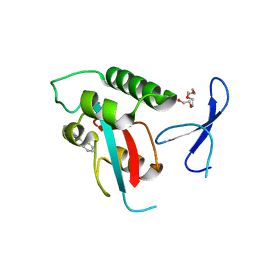 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-(MORPHOLIN-4-YLCARBONYL)-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2XCC
 
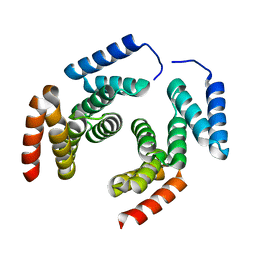 | | Crystal structure of PcrH from Pseudomonas aeruginosa | | Descriptor: | REGULATORY PROTEIN PCRH | | Authors: | Job, V, Mattei, P.-J, Lemaire, D, Attree, I, Dessen, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Chaperone Recognition of Type III Secretion System Minor Translocator Proteins.
J.Biol.Chem., 285, 2010
|
|
2XFN
 
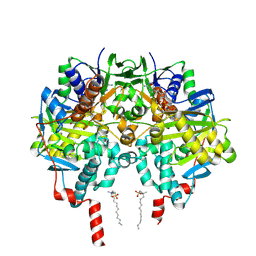 | | Human monoamine oxidase B in complex with 2-(2-benzofuranyl)-2- imidazoline | | Descriptor: | 2-(2-BENZOFURANYL)-2-IMIDAZOLINE, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bonivento, D, Milczek, E.M, McDonald, G.R, Binda, C, Holt, A, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potentiation of ligand binding through cooperative effects in monoamine oxidase B.
J. Biol. Chem., 285, 2010
|
|
3GW2
 
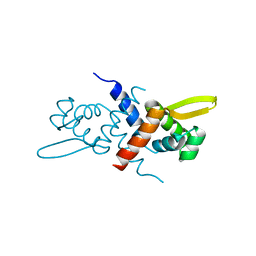 | | Crystal structure of possible transcriptional regulatory protein (fragment 1-100) from Mycobacterium bovis. Northeast Structural Genomics Consortium Target MbR242E. | | Descriptor: | Possible transcriptional regulatory arsR-family protein | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target MbR242E
To be published
|
|
3H6I
 
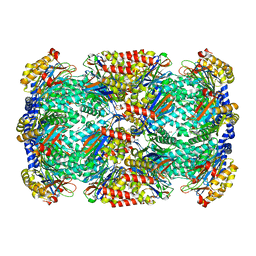 | |
2XHW
 
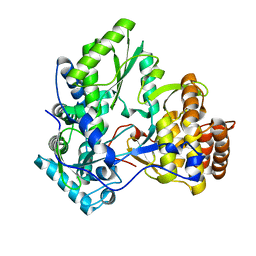 | | HCV-J4 NS5B Polymerase Trigonal Crystal Form | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
3H75
 
 | | Crystal Structure of a Periplasmic Sugar-binding protein from the Pseudomonas fluorescens | | Descriptor: | GLYCEROL, Periplasmic sugar-binding domain protein, SULFATE ION | | Authors: | Kumaran, D, Mahmood, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Periplasmic Sugar-binding protein from the Pseudomonas fluorescens
To be Published
|
|
3H7D
 
 | | The crystal structure of the cathepsin K Variant M5 in complex with chondroitin-4-sulfate | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Cathepsin K, ... | | Authors: | Cherney, M.M, Kienetz, M, Bromme, D, James, M.N.G. | | Deposit date: | 2009-04-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structure-activity analysis of cathepsin K/chondroitin 4-sulfate interactions.
J.Biol.Chem., 286, 2011
|
|
2XPC
 
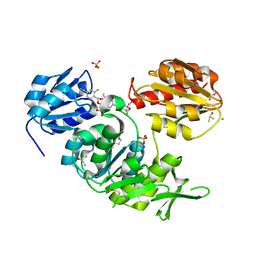 | | Second-generation sulfonamide inhibitors of MurD: Activity optimisation with conformationally rigid analogues of D-glutamic acid | | Descriptor: | (1R,3R,4S)-4-[({6-[(4-CYANO-2-FLUOROBENZYL)OXY]NAPHTHALEN-2-YL}SULFONYL)AMINO]CYCLOHEXANE-1,3-DICARBOXYLIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sosic, I, Barreteau, H, Simcic, M, Sink, R, Cesar, J, Golic-Grdadolnik, S, Contreras-Martel, C, Dessen, A, Amoroso, A, Joris, B, Blanot, D, Gobec, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Second-Generation Sulfonamide Inhibitors of D- Glutamic Acid-Adding Enzyme: Activity Optimisation with Conformationally Rigid Analogues of D- Glutamic Acid.
Eur.J.Med.Chem, 46, 2011
|
|
2XQQ
 
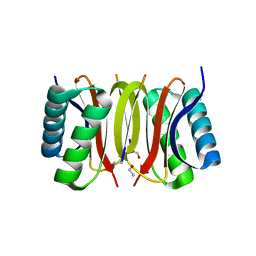 | | Human dynein light chain (DYNLL2) in complex with an in vitro evolved peptide (Ac-SRGTQTE). | | Descriptor: | ACETATE ION, DYNEIN LIGHT CHAIN 2, CYTOPLASMIC, ... | | Authors: | Rapali, P, Radnai, L, Suveges, D, Hetenyi, C, Harmat, V, Tolgyesi, F, Wahlgren, W.Y, Katona, G, Nyitray, L, Pal, G. | | Deposit date: | 2010-09-07 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Directed Evolution Reveals the Binding Motif Preference of the Lc8/Dynll Hub Protein and Predicts Large Numbers of Novel Binders in the Human Proteome
Plos One, 6, 2011
|
|
3HC0
 
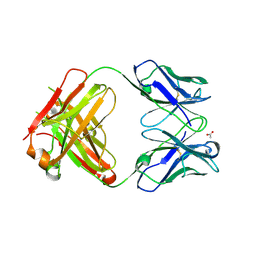 | | BHA10 IgG1 wild-type Fab - antibody directed at human LTBR | | Descriptor: | ACETATE ION, IMMUNOGLOBULIN IGG1 FAB, HEAVY CHAIN, ... | | Authors: | Arndt, J.W, Jordan, J.L, Lugovskoy, A, Wang, D. | | Deposit date: | 2009-05-05 | | Release date: | 2009-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural understanding of stabilization patterns in engineered bispecific Ig-like antibody molecules
Proteins, 77, 2009
|
|
3HCL
 
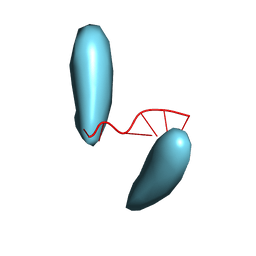 | | Helical superstructures in a DNA oligonucleotide crystal | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*AP*T)-3') | | Authors: | De Luchi, D, Martinez de Ilarduya, I, Subirana, J.A, Uson, I, Campos, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A geometric approach to the crystallographic solution of nonconventional DNA structures: helical superstructures of d(CGATAT)
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2XWA
 
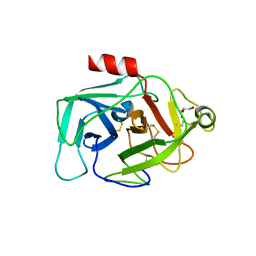 | | Crystal Structure of Complement Factor D Mutant R202A | | Descriptor: | COMPLEMENT FACTOR D, GLYCEROL | | Authors: | Forneris, F, Ricklin, D, Wu, J, Tzekou, A, Wallace, R.S, Lambris, J.D, Gros, P. | | Deposit date: | 2010-11-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of C3B in Complex with Factors B and D Give Insight Into Complement Convertase Formation.
Science, 330, 2010
|
|
3GSQ
 
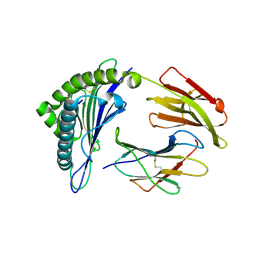 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-M5S peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant M5S (NLVPSVATV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
2XP9
 
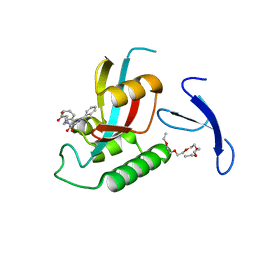 | | DISCOVERY OF CELL-ACTIVE PHENYL-IMIDAZOLE PIN1 INHIBITORS BY STRUCTURE-GUIDED FRAGMENT EVOLUTION | | Descriptor: | 4-[BENZYL(CARBOXYMETHYL)CARBAMOYL]-2-PHENYL-1H-IMIDAZOLE-5-CARBOXYLIC ACID, DODECAETHYLENE GLYCOL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE NIMA-INTERACTING 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-25 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Cell-Active Phenyl-Imidazole Pin1 Inhibitors by Structure-Guided Fragment Evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2Y0C
 
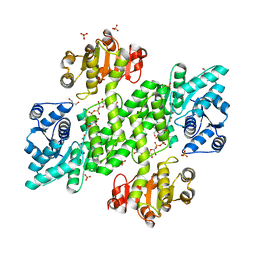 | | BceC mutation Y10S | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Rocha, J, Popescu, A.O, Borges, P, Mil-Homens, D, Sa-Correia, I, Fialho, A.M, Frazao, C. | | Deposit date: | 2010-12-02 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Burkholderia Cepacia Udp-Glucose Dehydrogenase (Ugd) Bcec and Role of Tyr10 in Final Hydrolysis of Ugd Thioester Intermediate.
J.Bacteriol., 193, 2011
|
|
3GTG
 
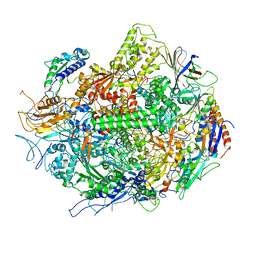 | | Backtracked RNA polymerase II complex with 12mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
