2JS5
 
 | | NMR Structure of protein Q60C73_METCA. Northeast Structural Genomics Consortium target McR1 | | Descriptor: | Uncharacterized protein | | Authors: | Singarapu, K.K, Wu, Y, Eletsky, A, Sukumaran, D, Parish, D, Chen, C.X, Nwosu, C, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of protein Q60C73_METCA.
To be Published
|
|
1BZO
 
 | | THREE-DIMENSIONAL STRUCTURE OF PROKARYOTIC CU,ZN SUPEROXIDE DISMUTASE FROM P.LEIOGNATHI, SOLVED BY X-RAY CRYSTALLOGRAPHY. | | Descriptor: | COPPER (II) ION, PROTEIN (SUPEROXIDE DISMUTASE), URANYL (VI) ION, ... | | Authors: | Bordo, D, Matak, D, Djinovic-Carugo, K, Rosano, C, Pesce, A, Bolognesi, M, Stroppolo, M.E, Falconi, M, Battistoni, A, Desideri, A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolutionary constraints for dimer formation in prokaryotic Cu,Zn superoxide dismutase.
J.Mol.Biol., 285, 1999
|
|
2EXH
 
 | | Structure of the family43 beta-Xylosidase from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Yuval, S, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EZ2
 
 | | Apo tyrosine phenol-lyase from Citrobacter freundii at pH 8.0 | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, Tyrosine phenol-lyase | | Authors: | Milic, D, Matkovic-Calogovic, D, Demidkina, T.V, Antson, A.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of apo- and holo-tyrosine phenol-lyase reveal a catalytically critical closed conformation and suggest a mechanism for activation by K+ ions
Biochemistry, 45, 2006
|
|
1GJ7
 
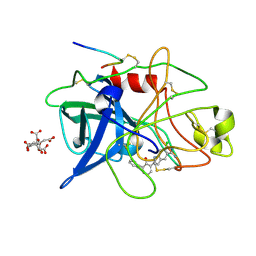 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJ9
 
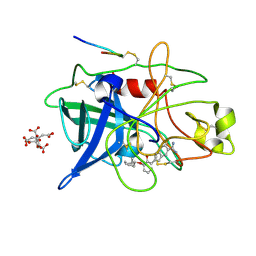 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 6-FLUORO-2-[2-HYDROXY-3-(2-METHYL-CYCLOHEXYLOXY)-PHENYL]-1H-INDOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-30 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
2JF5
 
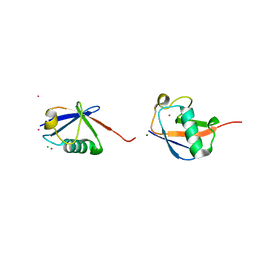 | | crystal structure of Lys63-linked di-ubiquitin | | Descriptor: | CADMIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Komander, D, Odenwaelder, P, Barford, D. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Discrimination of Structurally Equivalent Lys 63-Linked and Linear Polyubiquitin Chains.
Embo Rep., 10, 2009
|
|
1GJ6
 
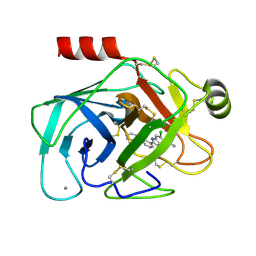 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, BETA-TRYPSIN, CALCIUM ION | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJ8
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 6-FLUORO-2-(2-HYDROXY-3-ISOBUTOXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJA
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | CITRIC ACID, N-(4-CARBAMIMIDOYL-PHENYL)-2-HYDROXY-BENZAMIDE, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
1GJ5
 
 | | SELECTIVITY AT S1, H2O DISPLACEMENT, UPA, TPA, SER190/ALA190 PROTEASE, STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 2-(2-HYDROXY-BIPHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, ACETYL HIRUDIN, SODIUM ION, ... | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
4R80
 
 | | Crystal Structure of a De Novo Designed Beta Sheet Protein, Cystatin Fold, Northeast Structural Genomics Consortium (NESG) Target OR486 | | Descriptor: | OR486 | | Authors: | Guan, R, Marcos, E, O'Connell, P, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Crystal Structure of an engineered protein with denovo beta sheet design, Northeast Structural Genomics Consortium (NESG) Target OR486
To be Published
|
|
5TQW
 
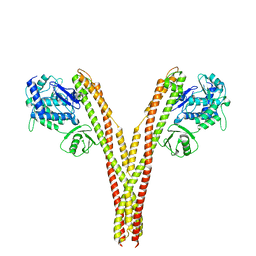 | | CryoEM reconstruction of human IKK1, open conformation 1 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswath, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
6BW3
 
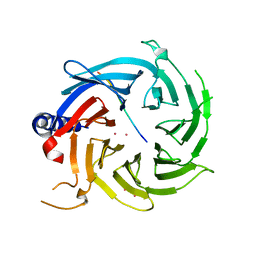 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
6BW4
 
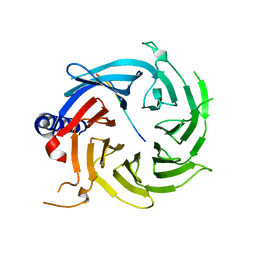 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
2K5G
 
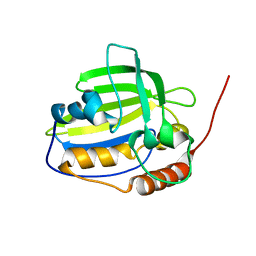 | | Solution NMR structure of protein encoded by gene BPP1335 from Bordetella parapertussis: Northeast Structural Genomics Target BpR195 | | Descriptor: | uncharacterized protein | | Authors: | Singarapu, K, Eletsky, A, Sathyamoorthy, B, Sukumaran, D, Wang, D, Jiang, M, Ciccosanti, C, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-27 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein encoded by gene BPP1335 from
Bordetella parapertussis: Northeast Structural Genomics Target BpR195
To be Published
|
|
4RAB
 
 | | Aza-acyclic nucleoside phosphonates containing a second phosphonate group as inhibitors of the human, Plasmodium falciparum and vivax 6-oxopurine phosphoribosyltransferases and their pro-drugs as antimalarial agents | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Keough, D.T, Hockov, D, Janeba, Z, Wang, T.-H, Naesens, L, Edstein, M.D, Chavchich, M, Guddat, L.W. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.264 Å) | | Cite: | Aza-acyclic Nucleoside Phosphonates Containing a Second Phosphonate Group As Inhibitors of the Human, Plasmodium falciparum and vivax 6-Oxopurine Phosphoribosyltransferases and Their Prodrugs As Antimalarial Agents.
J.Med.Chem., 58, 2015
|
|
2KAT
 
 | | Solution structure of protein BPP2914 from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR206 | | Descriptor: | uncharacterized protein | | Authors: | Wu, Y, Mills, J.L, Wang, D, Jiang, M, Foote, E.L, Xiao, R, Sathyamoorthy, B, Sukumaran, D, Nair, R, Everett, J.K, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein BPP2914 from Bordetella parapertussis. Northeast Structural Genomics Consortium target BpR206
To be Published
|
|
4RF3
 
 | | Crystal Structure of ketoreductase from Lactobacillus kefir, mutant A94F | | Descriptor: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | Authors: | Tang, Y, Tibrewal, N, Cascio, D. | | Deposit date: | 2014-09-24 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1SQP
 
 | | Crystal Structure Analysis of Bovine Bc1 with Myxothiazol | | Descriptor: | (2Z,6E)-7-{2'-[(2E,4E)-1,6-DIMETHYLHEPTA-2,4-DIENYL]-2,4'-BI-1,3-THIAZOL-4-YL}-3,5-DIMETHOXY-4-METHYLHEPTA-2,6-DIENAMID E, (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-19 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
4QRG
 
 | |
4QRV
 
 | | Crystal structure of I86F mutant of papain | | Descriptor: | CHLORIDE ION, Papain, SODIUM ION | | Authors: | Dutta, S, Choudhury, D, Roy, S. | | Deposit date: | 2014-07-02 | | Release date: | 2015-08-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Pro-peptide regulates the substrate specificity and zymogen activation process of papain: A structural and mechanistic insight
to be published
|
|
1MS6
 
 | | Dipeptide Nitrile Inhibitor Bound to Cathepsin S. | | Descriptor: | Cathepsin S, MORPHOLINE-4-CARBOXYLIC ACID [1S-(2-BENZYLOXY-1R-CYANO-ETHYLCARBAMOYL)-3-METHYL-BUTYL]AMIDE | | Authors: | Ward, Y.D, Thomson, D.S, Frye, L.L, Cywin, C.L, Morwick, T, Emmanuel, M.J, Zindell, R, McNeil, D, Bekkali, Y, Giradot, M, Hrapchak, M, DeTuri, M, Crane, K, White, D, Pav, S, Wang, Y, Hao, M.H, Grygon, C.A, Labadia, M.E, Freeman, D.M, Davidson, W, Hopkins, J.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of dipeptide nitriles as reversible and potent Cathepsin S inhibitors
J.Med.Chem., 45, 2002
|
|
1MUI
 
 | | Crystal structure of HIV-1 protease complexed with Lopinavir. | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, protease | | Authors: | Stoll, V, Qin, W, Stewart, K.D, Jakob, C, Park, C, Walter, K, Simmer, R.L, Helfrich, R, Bussiere, D, Kao, J, Kempf, D, Sham, H.L, Norbeck, D.W. | | Deposit date: | 2002-09-23 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Structure of ABT-378 (Lopinavir) Bound to HIV-1 Protease
BIOORG.MED.CHEM., 10, 2002
|
|
6CN2
 
 | | Crystal structure of zebrafish Phosphatidylinositol-4-phosphate 5- kinase alpha isoform D236N with bound ATP/Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Phosphatidylinositol-4-phosphate 5-kinase, ... | | Authors: | Zeng, X, Sui, D, Hu, J. | | Deposit date: | 2018-03-07 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural insights into lethal contractural syndrome type 3 (LCCS3) caused by a missense mutation of PIP5K gamma.
Biochem. J., 475, 2018
|
|
