6R6V
 
 | | Structure of recombinant human butyrylcholinesterase in complex with a fluorescent coumarin-based probe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cholinesterase, ... | | Authors: | Brazzolotto, X, Nachon, F, Knez, D, Gobec, S. | | Deposit date: | 2019-03-28 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of potent reversible selective inhibitors of butyrylcholinesterase as fluorescent probes.
J Enzyme Inhib Med Chem, 35, 2020
|
|
5Q0C
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with fructose-2,6-diphosphate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-(4-methoxyphenyl)thiophene-2-sulfonamide | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(5-bromo-1,3-thiazol-2-yl)-3-[5-(4-methoxyphenyl)thiophen-2-yl]sulfonylurea and with f2,6p
To be published
|
|
3MWJ
 
 | |
6TLA
 
 | | CRYSTAL STRUCTURE OF LECTIN-LIKE OX-LDL RECEPTOR 1 (C 1 2 1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Oxidized low-density lipoprotein receptor 1 | | Authors: | Nar, H, Fiegen, D, Schnapp, G. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A small-molecule inhibitor of lectin-like oxidized LDL receptor-1 acts by stabilizing an inactive receptor tetramer state
Commun Chem, 2020
|
|
1E6N
 
 | | Chitinase B from Serratia marcescens inactive mutant E144Q in complex with N-acetylglucosamine-pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE B, GLYCEROL, ... | | Authors: | Komander, D, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2000-08-21 | | Release date: | 2001-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of a Family 18 Exo-Chitinase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1E4X
 
 | | crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment | | Descriptor: | CYCLIC PEPTIDE, TAB2 | | Authors: | Hahn, M, Winkler, D, Misselwitz, R, Wessner, H, Welfle, K, Zahn, G, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Reactive Binding of Cyclic Peptides to an Anti-Tgf Alpha Antibody Fab Fragment: An X-Ray Structural and Thermodynamic Analysis
J.Mol.Biol., 314, 2001
|
|
1MIZ
 
 | | Crystal structure of an integrin beta3-talin chimera | | Descriptor: | TALIN, integrin beta3 | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
1M32
 
 | | Crystal Structure of 2-aminoethylphosphonate Transaminase | | Descriptor: | 2-aminoethylphosphonate-pyruvate aminotransferase, PHOSPHATE ION, PHOSPHONOACETALDEHYDE, ... | | Authors: | Chen, C.C.H, Zhang, H, Kim, A.D, Howard, A, Sheldrick, G.M, Mariano-Dunnaway, D, Herzberg, O. | | Deposit date: | 2002-06-26 | | Release date: | 2002-11-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Degradation Pathway of the Phosphonate Ciliatine: Crystal Structure of 2-Aminoethylphosphonate Transaminase
Biochemistry, 41, 2002
|
|
3N2K
 
 | | TUBULIN-NSC 613862: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Barbier, P, Dorleans, A, Devred, F, Sanz, L, Allegro, D, Alfonso, C, Knossow, M, Peyrot, V, Andreu, J.M. | | Deposit date: | 2010-05-18 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers.
J.Biol.Chem., 285, 2010
|
|
6RAG
 
 | | Heterodimeric ABC exporter TmrAB in inward-facing wide conformation under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RAN
 
 | | Heterodimeric ABC exporter TmrAB in inward-facing wide conformation | | Descriptor: | Multidrug resistance ABC transporter ATP-binding and permease protein, Nanobody Nb9F10 | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
6RBF
 
 | | Mucin 2 D3 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2019-04-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Intestinal Gel-Forming Mucins Polymerize by Disulfide-Mediated Dimerization of D3 Domains.
J.Mol.Biol., 431, 2019
|
|
6U0M
 
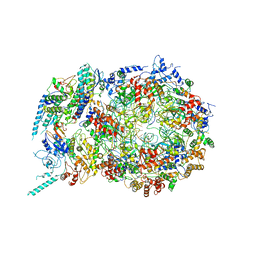 | | Structure of the S. cerevisiae replicative helicase CMG in complex with a forked DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (15-MER), ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Zhang, D, O'Donnell, M, Li, H. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA unwinding mechanism of a eukaryotic replicative CMG helicase.
Nat Commun, 11, 2020
|
|
6R30
 
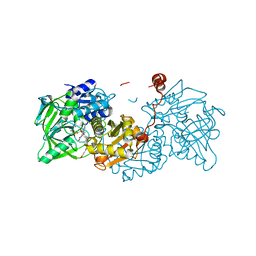 | |
6R3G
 
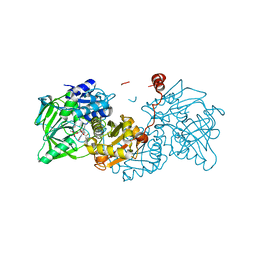 | |
6U3F
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Panton-Valentine Leucocidin F, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
1MOJ
 
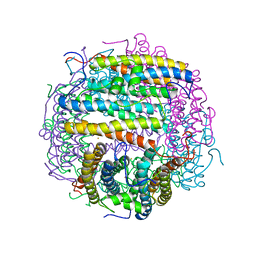 | | Crystal structure of an archaeal dps-homologue from Halobacterium salinarum | | Descriptor: | Dps-like ferritin, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Zeth, K, Offermann, S, Essen, L.O, Oesterhelt, D. | | Deposit date: | 2002-09-09 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron-oxo clusters biomineralizing on protein surfaces: Structural analysis of Halobacterium salinarum DpsA in its low- and high-iron states.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1EUB
 
 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN COLLAGENASE-3 (MMP-13) COMPLEXED TO A POTENT NON-PEPTIDIC SULFONAMIDE INHIBITOR | | Descriptor: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | Authors: | Zhang, X, Gonnella, N.C, Koehn, J, Pathak, N, Ganu, V, Melton, R, Parker, D, Hu, S.I, Nam, K.Y. | | Deposit date: | 2000-04-14 | | Release date: | 2001-04-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of human collagenase-3 (MMP-13) complexed to a potent non-peptidic sulfonamide inhibitor: binding comparison with stromelysin-1 and collagenase-1.
J.Mol.Biol., 301, 2000
|
|
6E9C
 
 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
3N30
 
 | | Crystal Structure of cubic Zn3-hUb (human ubiquitin) adduct | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Siliqi, D, Caliandro, R, Arnesano, F, Natile, G, Falini, G, Fermani, S, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
6E9T
 
 | | DHF58 filament | | Descriptor: | DHF58 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6RW3
 
 | | The molecular basis for sugar import in malaria parasites. | | Descriptor: | Hexose transporter 1, alpha-D-glucopyranose, beta-D-glucopyranose | | Authors: | Qureshi, A, Matsuoka, R, Brock, J, Drew, D. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The molecular basis for sugar import in malaria parasites.
Nature, 578, 2020
|
|
6U2S
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Bi-component leukocidin LukED subunit D, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6EOC
 
 | | Crystal structure of AMPylated GRP78 in apo form (Crystal form 2) | | Descriptor: | 78 kDa glucose-regulated protein, CITRATE ANION, SULFATE ION | | Authors: | Yan, Y, Preissler, S, Ron, D, Read, R.J. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
1M1P
 
 | | P21 crystal structure of the tetraheme cytochrome c3 from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, Small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
