1MDW
 
 | | Crystal Structure of Calcium-Bound Protease Core of Calpain II Reveals the Basis for Intrinsic Inactivation | | Descriptor: | CALCIUM ION, Calpain II, catalytic subunit | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Jia, Z, Davies, P.L. | | Deposit date: | 2002-08-07 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Calpain silencing by a reversible intrinsic mechanism.
Nat.Struct.Biol., 10, 2003
|
|
1MI0
 
 | | Crystal Structure of the redesigned protein G variant NuG2 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Biochemistry, 11, 2002
|
|
1MZN
 
 | | CRYSTAL STRUCTURE at 1.9 ANGSTROEMS RESOLUTION OF THE HOMODIMER OF HUMAN RXR ALPHA LIGAND BINDING DOMAIN BOUND TO THE SYNTHETIC AGONIST COMPOUND BMS 649 AND A COACTIVATOR PEPTIDE | | Descriptor: | 4-[2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-[1,3]DIOXOLAN-2-YL]-BENZOIC ACID, Nuclear receptor coactivator 2, RXR retinoid X receptor | | Authors: | Egea, P.F, Mitschler, A, Moras, D. | | Deposit date: | 2002-10-09 | | Release date: | 2002-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Recognition of Agonist Ligands by RXRs
MOL.ENDOCRINOL., 16, 2002
|
|
4V84
 
 | | Crystal structure of a complex containing domain 3 of CrPV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8RKH
 
 | | Crystal structure of the ZP-N2 and ZP-N3 domains of mouse ZP2 (mZP2-N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, ... | | Authors: | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKI
 
 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | Descriptor: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | Authors: | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RKE
 
 | | Crystal structure of the complete N-terminal region of human ZP2 (hZP2-N1N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
5VL4
 
 | | Accidental minimum contact crystal lattice formed by a redesigned protein oligomer | | Descriptor: | T33-53H-B | | Authors: | Cannon, K.A, Cascio, D, Sawaya, M.R, Park, R, Boyken, S, King, N, Yeates, T. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Design and structure of two new protein cages illustrate successes and ongoing challenges in protein engineering.
Protein Sci., 29, 2020
|
|
7LMX
 
 | | A HIGHLY SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 WITH A DISULFIDE | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7LMV
 
 | | SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
6DHP
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.04 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
3UMK
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with copper | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
3UID
 
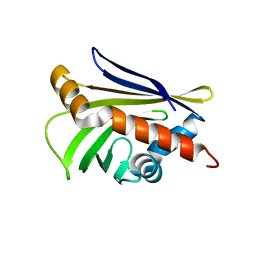 | | Crystal Structure of Protein Ms6760 from Mycobacterium smegmatis | | Descriptor: | Putative uncharacterized protein | | Authors: | Bajaj, R.A, Miallau, L, Cascio, D, Arbing, M, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-11-04 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Crystal structure of the toxin Msmeg_6760, the structural homolog of Mycobacterium tuberculosis Rv2035, a novel type II toxin involved in the hypoxic response.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
3UMH
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with cadmium | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
5KO0
 
 | | Human Islet Amyloid Polypeptide Segment 15-FLVHSSNNFGA-25 Determined by MicroED | | Descriptor: | THIOCYANATE ION, hIAPP(15-25)WT | | Authors: | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.4 Å) | | Cite: | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
1VDY
 
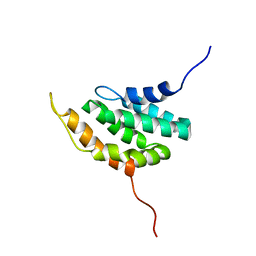 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | Descriptor: | hypothetical protein (RAFL09-17-B18) | | Authors: | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
8DYA
 
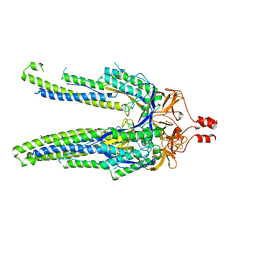 | |
1W9A
 
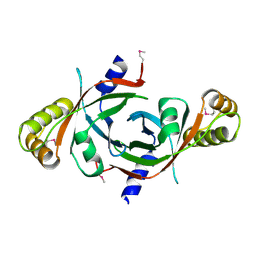 | | Crystal structure of Rv1155 from Mycobacterium tuberculosis | | Descriptor: | PUTATIVE PYRIDOXINE/PYRIDOXAMINE 5'-PHOSPHATE OXIDASE | | Authors: | Cannan, S, Sulzenbacher, G, Roig-Zamboni, V, Scappuccini, L, Frassinetti, F, Maurien, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2004-10-07 | | Release date: | 2005-01-06 | | Last modified: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Conserved Hypothetical Protein Rv1155 from Mycobacterium Tuberculosis
FEBS Lett., 579, 2005
|
|
6Z3Z
 
 | |
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7Y38
 
 | |
4U01
 
 | | HCV NS3/4A serine protease in complex with 6570 | | Descriptor: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | Authors: | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
8ERR
 
 | |
