1ZII
 
 | |
7QUF
 
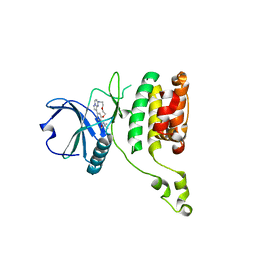 | | The STK17A (DRAK1) Kinase Domain Bound to CK156 | | Descriptor: | Serine/threonine-protein kinase 17A, ~{N}-~{tert}-butyl-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Dederer, V, Kurz, C, Hanke, T, Knapp, S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
J.Med.Chem., 65, 2022
|
|
7ZY3
 
 | | Room temperature structure of Archaerhodopsin-3 obtained 110 ns after photoexcitation | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kwan, T.O.C, Judge, P.J, Moraes, I, Watts, A, Axford, D, Bada Juarez, J.F. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A versatile approach to high-density microcrystals in lipidic cubic phase for room-temperature serial crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
6TSO
 
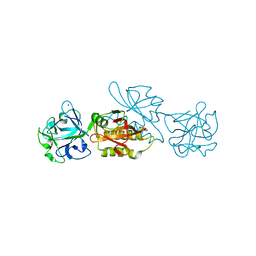 | | Marasmius oreades agglutinin (MOA) inhibited by cadmium | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CADMIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
6TSW
 
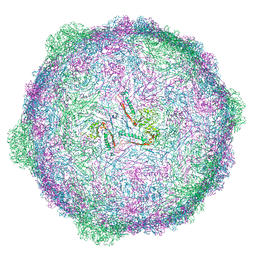 | | Isometric capsid of empty GTA particle computed with I4(I,n25r) symmetry | | Descriptor: | Major capsid protein Rcc01687 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
1YMG
 
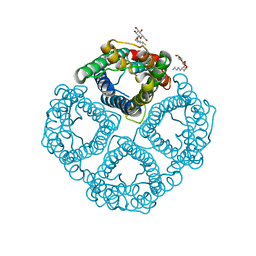 | | The Channel Architecture of Aquaporin O at 2.2 Angstrom Resolution | | Descriptor: | Lens fiber major intrinsic protein, nonyl beta-D-glucopyranoside | | Authors: | Harries, W.E.C, Akhavan, D, Miercke, L.J.W, Khademi, S, Stroud, R.M. | | Deposit date: | 2005-01-20 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Channel Architecture of Aquaporin 0 at a 2.2-A Resolution
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8A4V
 
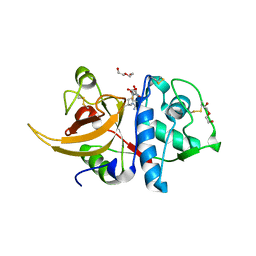 | | Crystal structure of human cathepsin L with covalently bound E-64 | | Descriptor: | Cathepsin L, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
2V9F
 
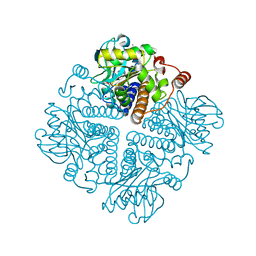 | |
6TU1
 
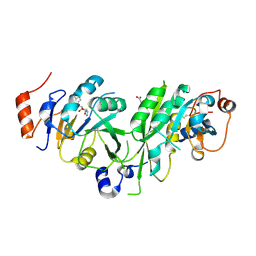 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 8 (ASI_M3M_091) | | Descriptor: | (2~{R},3~{R},4~{R},5~{R})-2-[(6-aminopurin-9-yl)methyl]-5-azanyl-oxane-3,4-diol, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-12-31 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer.
Chemmedchem, 15, 2020
|
|
1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | Authors: | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
5L33
 
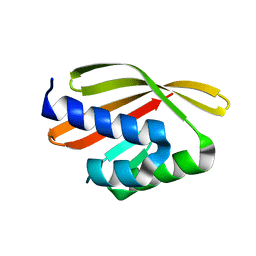 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | denovo NTF2 | | Authors: | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-08-03 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
4V2W
 
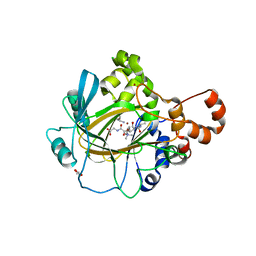 | | JMJD2A COMPLEXED WITH NI(II), NOG AND HISTONE H3K27me3 PEPTIDE (16-35) | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE H3.1T, ... | | Authors: | Chowdhury, R, Zafred, D, Schofield, C.J. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Studies on the Catalytic Domains of Multiple Jmjc Oxygenases Using Peptide Substrates.
Epigenetics, 9, 2014
|
|
2UY6
 
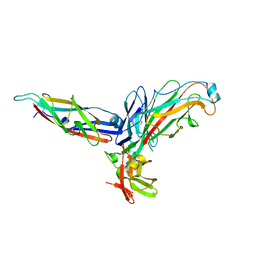 | | Crystal structure of the P pilus rod subunit PapA | | Descriptor: | PAP FIMBRIAL MAJOR PILIN PROTEIN, PERIPLASMID CHAPERONE PAPD PROTEIN | | Authors: | Verger, D, Bullitt, E, Hultgren, S.J, Waksman, G. | | Deposit date: | 2007-04-02 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the P Pilus Rod Subunit Papa.
Plos Pathog., 3, 2007
|
|
8A4W
 
 | | Crystal structure of human cathepsin L with covalently bound Cathepsin L inhibitor IV | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
2J75
 
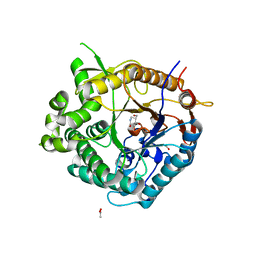 | | Beta-glucosidase from Thermotoga maritima in complex with noeuromycin | | Descriptor: | (2R,3S,4R,5R)-5-(HYDROXYMETHYL)PIPERIDINE-2,3,4-TRIOL, ACETATE ION, BETA-GLUCOSIDASE A, ... | | Authors: | Gloster, T.M, Meloncelli, P, Stick, R.V, Zechel, D, Davies, G.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
7QF4
 
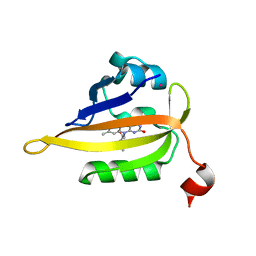 | | Structure of the R57Q mutant of miniSOG expressed in E. coli in LB medium enriched with riboflavin | | Descriptor: | CHLORIDE ION, COBALT (II) ION, RIBOFLAVIN, ... | | Authors: | Lafaye, C, Aumonier, S, von Stetten, D, Noirclerc-Savoye, N, Gotthard, G, Royant, A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Riboflavin-binding proteins for singlet oxygen production.
Photochem Photobiol Sci, 21, 2022
|
|
8A4X
 
 | | Crystal structure of human Cathepsin L with covalently bound Calpain inhibitor III | | Descriptor: | (phenylmethyl) N-[(2S)-3-methyl-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-phenyl-propan-2-yl]amino]butan-2-yl]carbamate, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Falke, S, Lieske, J, Guenther, S, Reinke, P.Y.A, Ewert, W, Loboda, J, Karnicar, K, Usenik, A, Lindic, N, Sekirnik, A, Chapman, H.N, Hinrichs, W, Turk, D, Meents, A. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Elucidation and Antiviral Activity of Covalent Cathepsin L Inhibitors.
J.Med.Chem., 67, 2024
|
|
1YM5
 
 | | Crystal structure of YHI9, the yeast member of the phenazine biosynthesis PhzF enzyme superfamily. | | Descriptor: | Hypothetical 32.6 kDa protein in DAP2-SLT2 intergenic region | | Authors: | Liger, D, Quevillon-Cheruel, S, Sorel, I, Bremang, M, Blondeau, K, Aboulfath, I, Janin, J, Van Tilbeurgh, H, Leulliot, N, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2005-01-20 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of YHI9, the yeast member of the phenazine biosynthesis PhzF enzyme superfamily
Proteins, 60, 2005
|
|
2VKG
 
 | | COMPLEXES OF DODECIN WITH FLAVIN AND FLAVIN-LIKE LIGANDS | | Descriptor: | CHLORIDE ION, DODECIN, MAGNESIUM ION, ... | | Authors: | Grininger, M, Noell, G, Trawoeger, S, Sinner, E, Oesterhelt, D. | | Deposit date: | 2007-12-19 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Electrochemical Switching of the Flavoprotein Dodecin at Gold Surfaces Modified by Flavin-DNA Hybrid Linkers
Biointerphases, 3, 2008
|
|
7QF2
 
 | | Structure of miniSOG reconstituted with riboflavin as a cofactor | | Descriptor: | CHLORIDE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Lafaye, C, Aumonier, S, von Stetten, D, Noirclerc-Savoye, N, Gotthard, G, Royant, A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Riboflavin-binding proteins for singlet oxygen production.
Photochem Photobiol Sci, 21, 2022
|
|
1YMT
 
 | | Mouse SF-1 LBD | | Descriptor: | 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL, Nuclear receptor 0B2, Steroidogenic factor 1 | | Authors: | Krylova, I.N, Sablin, E.P, Moore, J, Xu, R.X, Waitt, G.M, Juzumiene, D, Bynum, J.M, Fletterick, R.J, Willson, T.M, Ingraham, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
8CUB
 
 | |
7QF5
 
 | | Structure of the Q103L mutant of miniSOG | | Descriptor: | CHLORIDE ION, COBALT (II) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Lafaye, C, Aumonier, S, von Stetten, D, Noirclerc-Savoye, N, Gotthard, G, Royant, A. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Riboflavin-binding proteins for singlet oxygen production.
Photochem Photobiol Sci, 21, 2022
|
|
2V7T
 
 | |
2V65
 
 | | Apo LDH from the psychrophile C. gunnari | | Descriptor: | L-LACTATE DEHYDROGENASE A CHAIN | | Authors: | Coquelle, N, Madern, D, Vellieux, F. | | Deposit date: | 2007-07-13 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Activity, Stability and Structural Studies of Lactate Dehydrogenases Adapted to Extreme Thermal Environments.
J.Mol.Biol., 374, 2007
|
|
