3C5Q
 
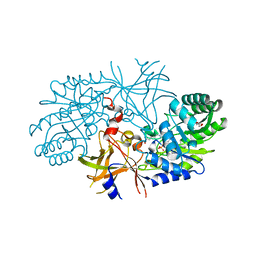 | | Crystal structure of diaminopimelate decarboxylase (I148L mutant) from Helicobacter pylori complexed with L-lysine | | Descriptor: | Diaminopimelate decarboxylase, GLYCEROL, LYSINE, ... | | Authors: | Hu, T, Wu, D, Jiang, H, Shen, X. | | Deposit date: | 2008-02-01 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of diaminopimelate decarboxylase from Helicobacter pylori
To be Published
|
|
3TCP
 
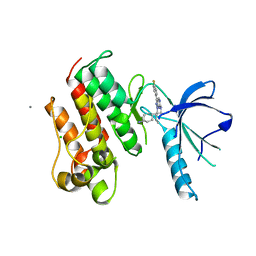 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | Descriptor: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
3C8A
 
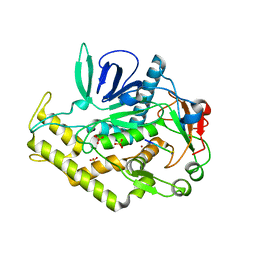 | |
3C90
 
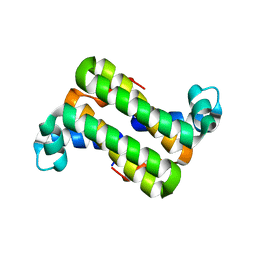 | | The 1.25 A Resolution Structure of Phosphoribosyl-ATP Pyrophosphohydrolase from Mycobacterium tuberculosis, crystal form II | | Descriptor: | Phosphoribosyl-ATP pyrophosphatase | | Authors: | Javid-Majd, F, Yang, D, Ioerger, T.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-02-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The 1.25 A resolution structure of phosphoribosyl-ATP pyrophosphohydrolase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
8OHI
 
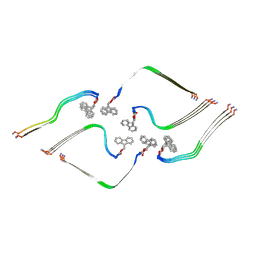 | | Structure of the Fmoc-Tau-PAM4 Type 2 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OH2
 
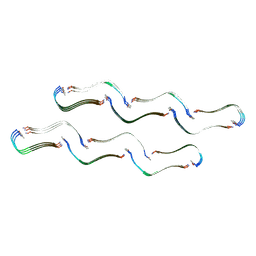 | | Structure of the Tau-PAM4 Type 1 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
3BKD
 
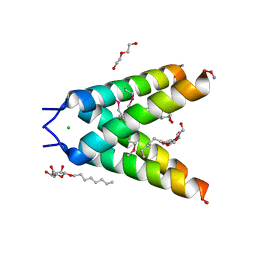 | | High resolution Crystal structure of Transmembrane domain of M2 protein | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Transmembrane Domain of Matrix protein M2, ... | | Authors: | Stouffer, A.L, Acharya, R, Salom, D. | | Deposit date: | 2007-12-06 | | Release date: | 2008-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for the function and inhibition of an influenza virus proton channel
Nature, 451, 2008
|
|
8OHP
 
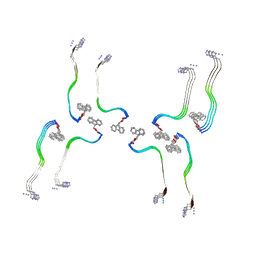 | | Structure of the Fmoc-Tau-PAM4 Type 3 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
8OI0
 
 | | Structure of the Fmoc-Tau-PAM4 Type 4 amyloid fibril | | Descriptor: | Microtubule-associated protein tau | | Authors: | Wilkinson, M, Louros, N, Tsaka, G, Ramakers, M, Morelli, C, Garcia, T, Gallardo, R.U, D'Haeyer, S, Goossens, V, Audenaert, D, Thal, D.R, Ranson, N.A, Radford, S.E, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Local structural preferences in shaping tau amyloid polymorphism.
Nat Commun, 15, 2024
|
|
3BYI
 
 | | Crystal structure of human Rho GTPase activating protein 15 (ARHGAP15) | | Descriptor: | Rho GTPase activating protein 15 | | Authors: | Shrestha, L, Tickle, J, Elkins, J, Burgess-Brown, N, Johansson, C, Papagrigoriou, E, Kavanagh, K, Pike, A.C.W, Ugochukwu, E, Uppenberg, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Human Rho GTPase Activating Protein 15 (ARHGAP15).
To be Published
|
|
3PAA
 
 | | Mechanism of inactivation of E. coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid (S-ADFA) pH 8.0 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminofuran-2-carboxylic acid, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Fu, M, Silverman, R.B, Ringe, D. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-01 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of inactivation of Escherichia coli aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-furancarboxylic acid .
Biochemistry, 49, 2010
|
|
1UOP
 
 | |
3BS4
 
 | | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide | | Descriptor: | Uncharacterized protein PH0321, Unknown peptide | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of uncharacterized protein PH0321 from Pyrococcus horikoshii in complex with an unknown peptide.
To be Published
|
|
8ONT
 
 | | Structure of Setaria italica NRAT in complex with a nanobody | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, NRAMP related aluminium transporter, Nanobody1 | | Authors: | Ramanadane, K, Liziczai, M, Markovic, D, Straub, M.S, Rosalen, G.T, Udovcic, A, Dutzler, R, Manatschal, C. | | Deposit date: | 2023-04-03 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural and functional properties of a plant NRAMP-related aluminum transporter.
Elife, 12, 2023
|
|
1UN6
 
 | | THE CRYSTAL STRUCTURE OF A ZINC FINGER - RNA COMPLEX REVEALS TWO MODES OF MOLECULAR RECOGNITION | | Descriptor: | 5S RIBOSOMAL RNA, MAGNESIUM ION, TRANSCRIPTION FACTOR IIIA, ... | | Authors: | Lu, D, Searles, M.A, Klug, A. | | Deposit date: | 2003-09-04 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Zinc-Finger-RNA Complex Reveals Two Modes of Molecular Recognition
Nature, 426, 2003
|
|
3C4S
 
 | | Crystal structure of the Ssl0352 protein from Synechocystis sp. Northeast Structural Genomics Consortium target SgR42 | | Descriptor: | Ssl0352 protein | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Wang, D, Maglaqui, M, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Ssl0352 protein from Synechocystis sp.
To be Published
|
|
3C97
 
 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | Descriptor: | Signal transduction histidine kinase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3BKM
 
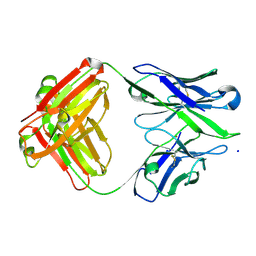 | | Structure of anti-amyloid-beta Fab WO2 (Form A, P212121) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa, ... | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
3FXU
 
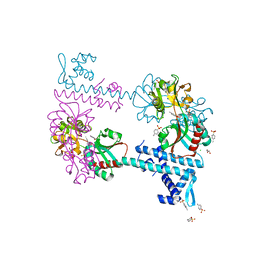 | | Crystal structure of TsaR in complex with its effector p-toluenesulfonate | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
2ZNJ
 
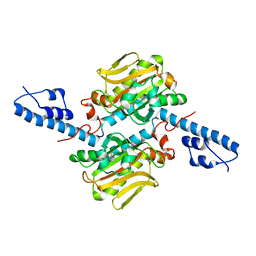 | | Crystal structure of Pyrrolysyl-tRNA synthetase from Desulfitobacterium hafniense | | Descriptor: | Putative uncharacterized protein | | Authors: | Nozawa, K, Araiso, Y, Soll, D, Ishitani, R, Nureki, O. | | Deposit date: | 2008-04-25 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality
Nature, 457, 2009
|
|
4RMK
 
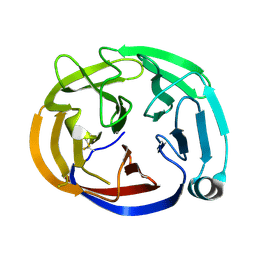 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in P65 crystal form | | Descriptor: | CALCIUM ION, Latrophilin-3 | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
2Z35
 
 | | Crystal structure of immune receptor | | Descriptor: | T-cell receptor alpha-chain, T-cell receptor beta-chain | | Authors: | Feng, D, Bond, C.J, Ely, L.K, Garcia, K.C. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'
Nat.Immunol., 8, 2007
|
|
2X8G
 
 | | Oxidized thioredoxin glutathione reductase from Schistosoma mansoni | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Angelucci, F, Dimastrogiovanni, D, Boumis, G, Brunori, M, Miele, A.E, Saccoccia, F, Bellelli, A. | | Deposit date: | 2010-03-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the Catalytic Cycle of Schistosoma Mansoni Thioredoxin Glutathione Reductase by X-Ray Crystallography
J.Biol.Chem., 285, 2010
|
|
2Z7J
 
 | | Structural insights into de multifunctional VP3 protein of birnaviruses:gold derivative | | Descriptor: | Capsid assembly protein VP3, GOLD ION | | Authors: | Casanas, A, Navarro, A, Ferrer-Orta, C, Gonzalez, D, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-08-23 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the multifunctional protein VP3 of birnaviruses.
Structure, 16, 2008
|
|
2Z90
 
 | | Crystal Structure of the Second Dps from Mycobacterium smegmatis | | Descriptor: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Roy, S, Saraswathi, R, Chatterji, D, Vijayan, M. | | Deposit date: | 2007-09-13 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies on the second Mycobacterium smegmatis Dps: invariant and variable features of structure, assembly and function.
J.Mol.Biol., 375, 2008
|
|
