7P9D
 
 | | Crystal structure of Chlamydomonas reinhardtii NADPH Dependent Thioredoxin Reductase 1 domain | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Singh, R.K, Marchetti, G.M, Hippler, M, Kuemmel, D. | | Deposit date: | 2021-07-27 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analysis revealed a novel conformation of the NTRC reductase domain from Chlamydomonas reinhardtii.
J.Struct.Biol., 214, 2021
|
|
4WFB
 
 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with BC-3205 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S rRNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4PWL
 
 | | Crystal structure of the complex between PPARgamma-LBD and the S enantiomer of Mbx-102 (Metaglidasen) | | Descriptor: | (2S)-(4-chlorophenyl)[3-(trifluoromethyl)phenoxy]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Lavecchia, A, Piemontese, L. | | Deposit date: | 2014-03-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | On the metabolically active form of metaglidasen: improved synthesis and investigation of its peculiar activity on peroxisome proliferator-activated receptors and skeletal muscles.
Chemmedchem, 10, 2015
|
|
7P9E
 
 | |
4WMF
 
 | | Crystal structure of catalytically inactive MERS-CoV 3CL protease (C148A) in spacegroup P212121 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MERS-CoV 3CL protease, TETRAETHYLENE GLYCOL | | Authors: | Lountos, G.T, Needle, D, Waugh, D.S. | | Deposit date: | 2014-10-08 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YQC
 
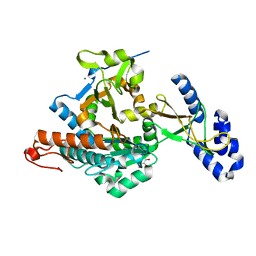 | | Crystal Structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the apo-like form | | Descriptor: | GLYCEROL, MAGNESIUM ION, UDP-N-acetylglucosamine pyrophosphorylase | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
6OEX
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor 3-(2-{1-[2-(Piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3- thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine | | Descriptor: | 3-(2-{1-[2-(piperidin-4-yl)ethyl]-1H-indol-5-yl}-5-[1-(pyrrolidin-1-yl)cyclohexyl]-1,3-thiazol-4-yl)-N-(2,2,2-trifluoroethyl)prop-2-yn-1-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
8SE7
 
 | |
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
4WF9
 
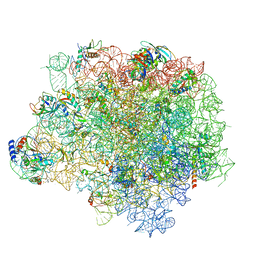 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with telithromycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A.E. | | Deposit date: | 2014-09-14 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.427 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8SDM
 
 | |
8SDP
 
 | |
3EHF
 
 | |
6OTC
 
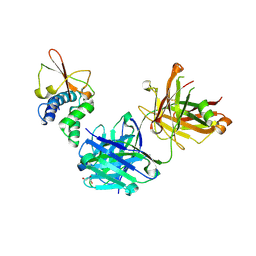 | | Synthetic Fab bound to Marburg virus VP35 interferon inhibitory domain | | Descriptor: | CHLORIDE ION, GLYCEROL, Polymerase cofactor VP35, ... | | Authors: | Amatya, P, Chen, G, Borek, D, Sidhu, S.S, Leung, D.W. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Marburg Virus RNA Synthesis by a Synthetic Anti-VP35 Antibody.
Acs Infect Dis., 5, 2019
|
|
3LIP
 
 | |
6O3V
 
 | | Crystal structure for RVA-VP3 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, Protein VP3, ... | | Authors: | Kumar, D, Yu, X, Wang, Z, Hu, L, Prasad, V. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2.7 angstrom cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity.
Sci Adv, 6, 2020
|
|
4X09
 
 | | Structure of human RNase 6 in complex with sulphate anions | | Descriptor: | GLYCEROL, Ribonuclease K6, SULFATE ION | | Authors: | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
8H8H
 
 | |
2Z60
 
 | | Crystal Structure of the T315I Mutant of Abl kinase bound with PPY-A | | Descriptor: | 5-[3-(2-METHOXYPHENYL)-1H-PYRROLO[2,3-B]PYRIDIN-5-YL]-N,N-DIMETHYLPYRIDINE-3-CARBOXAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Dalgarno, D, Zhu, X. | | Deposit date: | 2007-07-21 | | Release date: | 2007-09-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the T315I Mutant of Abl Kinase
Chem.Biol.Drug Des., 70, 2007
|
|
4GKQ
 
 | |
6OAO
 
 | | Structure of DBP in complex with human neutralizing antibody 092096 | | Descriptor: | Antibody 092096 single chain variable fragment, Duffy binding surface protein region II, SULFATE ION | | Authors: | Urusova, D, Tolia, N.H. | | Deposit date: | 2019-03-18 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.497 Å) | | Cite: | Structural basis for neutralization of Plasmodium vivax by naturally acquired human antibodies that target DBP.
Nat Microbiol, 4, 2019
|
|
8H5E
 
 | | Crystal structure of the MgtE TM domain in complex with Ca2+ ions at 2.5 angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, Magnesium transporter MgtE | | Authors: | Teng, X, Sheng, D, Hattori, M. | | Deposit date: | 2022-10-13 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ion selectivity mechanism of the MgtE channel for Mg 2+ over Ca 2 .
Iscience, 25, 2022
|
|
4X67
 
 | | Crystal structure of elongating yeast RNA polymerase II stalled at oxidative Cyclopurine DNA lesions. | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X6Z
 
 | | Yeast 20S proteasome in complex with PR-VI modulator | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rostankowski, R, Witkowska, J, Borek, D, Otwinowski, Z, Jankowska, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures revealed the common place of binding of low-molecular
mass activators with the 20S proteasome
To Be Published
|
|
1RCS
 
 | | NMR STUDY OF TRP REPRESSOR-OPERATOR DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*AP*CP*G)-3'), TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the trp repressor-operator DNA complex.
J.Mol.Biol., 238, 1994
|
|
