3NF5
 
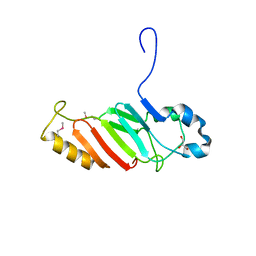 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
1Q1P
 
 | | E-Cadherin activation | | Descriptor: | CALCIUM ION, Epithelial-cadherin | | Authors: | Haussinger, D, Stetefeld, J. | | Deposit date: | 2003-07-22 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography.
Embo J., 23, 2004
|
|
1Q2J
 
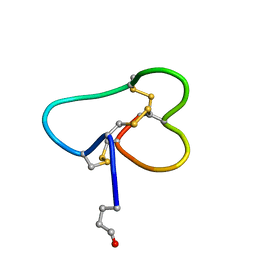 | | Structural basis for tetrodotoxin-resistant sodium channel binding by mu-conotoxin SmIIIA | | Descriptor: | Mu-conotoxin SmIIIA | | Authors: | Keizer, D.W, West, P.J, Lee, E.F, Olivera, B.M, Bulaj, G, Yoshikami, D, Norton, R.S. | | Deposit date: | 2003-07-24 | | Release date: | 2004-02-24 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structural basis for tetrodotoxin-resistant sodium channel binding by mu-conotoxin SmIIIA.
J.Biol.Chem., 278, 2003
|
|
1PL1
 
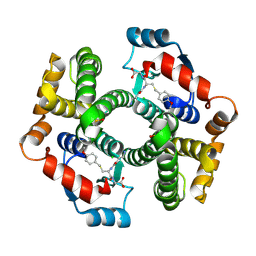 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with a decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1FVV
 
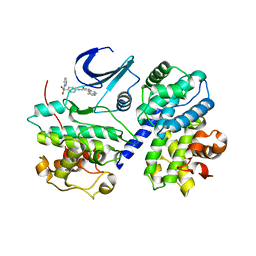 | | THE STRUCTURE OF CDK2/CYCLIN A IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(7-OXO-7H-THIAZOLO[5,4-E]INDOL-8-YLMETHYL)-AMINO]-N-PYRIDIN-2-YL-BENZENESULFONAMIDE, CYCLIN A, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
3NM2
 
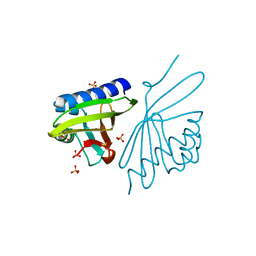 | | Crystal Structure of Ketosteroid Isomerase D38EP39GV40GS42G from Pseudomonas Testosteroni (tKSI) | | Descriptor: | SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38EP39GV40GS42G from Pseudomonas Testosteroni (tKSI)
To be Published
|
|
1PQD
 
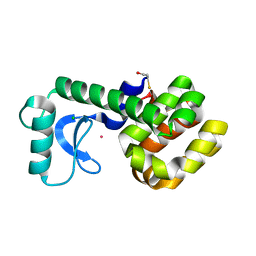 | | T4 LYSOZYME CORE REPACKING MUTANT CORE10/TA | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
6DJE
 
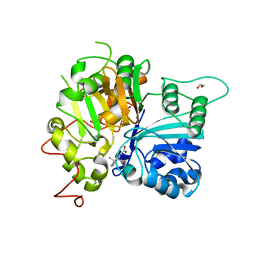 | | Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound CDS010292 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-(propan-2-yl)quinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
1PUX
 
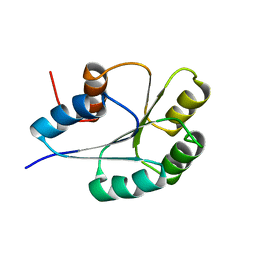 | | NMR Solution Structure of BeF3-Activated Spo0F, 20 conformers | | Descriptor: | Sporulation initiation phosphotransferase F | | Authors: | Gardino, A.K, Volkman, B.F, Cho, H.S, Lee, S.Y, Wemmer, D.E, Kern, D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of BeF(3)(-)-activated Spo0F reveals the conformational switch in a phosphorelay system.
J.Mol.Biol., 331, 2003
|
|
1FWO
 
 | | THE SOLUTION STRUCTURE OF A 35-RESIDUE FRAGMENT FROM THE GRANULIN/EPITHELIN-LIKE SUBDOMAIN OF RICE ORYZAIN BETA (ROB 382-416 (C398S,C399S,C407S,C413S)) | | Descriptor: | ORYZAIN BETA CHAIN | | Authors: | Tolkatchev, D, Xu, P, Ni, F. | | Deposit date: | 2000-09-24 | | Release date: | 2001-05-09 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | A peptide derived from the C-terminal part of a plant cysteine protease folds into a stack of two beta-hairpins, a scaffold present in the emerging family of granulin-like growth factors.
J.Pept.Res., 57, 2001
|
|
1FXJ
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-26 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
1PSY
 
 | | STRUCTURE OF RicC3, NMR, 20 STRUCTURES | | Descriptor: | 2S albumin | | Authors: | Pantoja-Uceda, D, Bruix, M, Gimenez-Gallego, G, Rico, M, Santoro, J. | | Deposit date: | 2003-06-22 | | Release date: | 2004-01-13 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis.
Biochemistry, 42, 2003
|
|
6VJK
 
 | | Streptavidin mutant M88 (N49C/A86C) | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Marangoni, J.M, Wu, S.C, Fogen, D, Wong, S.L, Ng, K.K.S. | | Deposit date: | 2020-01-16 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering a disulfide-gated switch in streptavidin enables reversible binding without sacrificing binding affinity.
Sci Rep, 10, 2020
|
|
6DLF
 
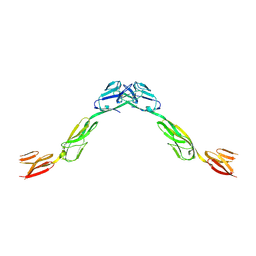 | | Crystal structure of NTRI homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Neurotrimin | | Authors: | Ranaivoson, F.M, Turk, L.S, Ozkan, E, Montelione, G.T, Comoletti, D. | | Deposit date: | 2018-06-01 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.446 Å) | | Cite: | Structure of a heterodimer of neuronal cell surface proteins
Structure, 2019
|
|
3NEP
 
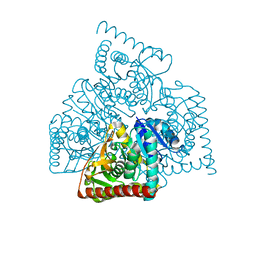 | |
1G25
 
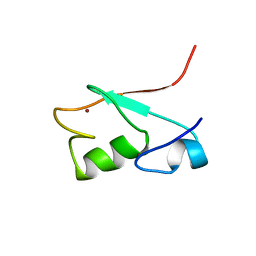 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF THE HUMAN TFIIH MAT1 SUBUNIT | | Descriptor: | CDK-ACTIVATING KINASE ASSEMBLY FACTOR MAT1, ZINC ION | | Authors: | Gervais, V, Wasielewski, E, Busso, D, Poterszman, A, Egly, J.M, Thierry, J.C, Kieffer, B. | | Deposit date: | 2000-10-17 | | Release date: | 2000-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the N-terminal Domain of the Human TFIIH MAT1 Subunit: New Insights into the RING Finger Family
J.Biol.Chem., 276, 2001
|
|
1G0O
 
 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND PYROQUILON | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYROQUILON, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Liao, D, Basarab, G.S, Gatenby, A.A, Valent, B, Jordan, D.B. | | Deposit date: | 2000-10-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of trihydroxynaphthalene reductase-fungicide complexes: implications for structure-based design and catalysis.
Structure, 9, 2001
|
|
3NFF
 
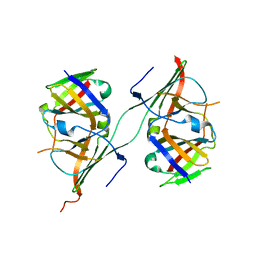 | | Crystal structure of extended Dimerization module of RNA polymerase I subcomplex A49/A34.5 | | Descriptor: | RNA polymerase I subunit A34.5, RNA polymerase I subunit A49 | | Authors: | Geiger, S.R, Lorenzen, K, Schreieck, A, Hanecker, P, Kostrewa, D, Heck, A.J.R, Cramer, P. | | Deposit date: | 2010-06-10 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex.
Mol.Cell, 39, 2010
|
|
1G1F
 
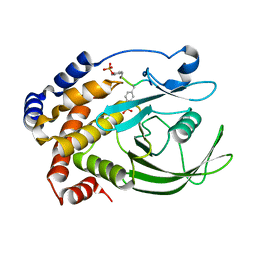 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH A TRI-PHOSPHORYLATED PEPTIDE (RDI(PTR)ETD(PTR)(PTR)RK) FROM THE INSULIN RECEPTOR KINASE | | Descriptor: | PROTEIN TYROSINE PHOSPHATASE 1B, TRI-PHOSPHORYLATED PEPTIDE FROM THE INSULIN RECEPTOR KINASE | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Tonks, N.K, Barford, D. | | Deposit date: | 2000-10-11 | | Release date: | 2001-01-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Mol.Cell, 6, 2000
|
|
1PXN
 
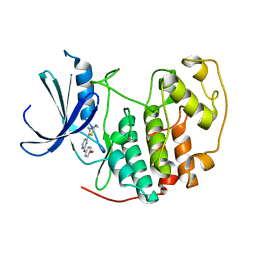 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-[4-(4-Methyl-2-methylamino-thiazol-5-yl)-pyrimidin-2-ylamino]-phenol | | Descriptor: | 4-[4-(4-METHYL-2-METHYLAMINO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-PHENOL, Cell division protein kinase 2 | | Authors: | Wang, S, Meades, C, Wood, G, Osnowski, A, Anderson, S, Yuill, R, Thomas, M, Mezna, M, Jackson, W, Midgley, C, Griffiths, G, McNae, I, Wu, S.Y, McInnes, C, Zheleva, D, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2003-07-04 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Anilino-4-(thiazol-5-yl)pyrimidine CDK inhibitors: synthesis, SAR analysis, X-ray crystallography, and biological activity.
J.Med.Chem., 47, 2004
|
|
1PZR
 
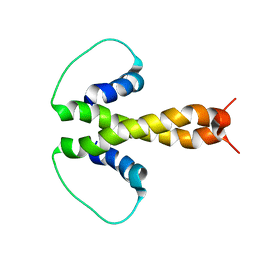 | | Structure of fused docking domains from the erythromycin polyketide synthase (DEBS), a model for the interaction between DEBS2 and DEBS3: the B domain | | Descriptor: | Erythronolide synthase | | Authors: | Broadhurst, R.W, Nietlispach, D, Wheatcroft, M.P, Leadlay, P.F, Weissman, K.J. | | Deposit date: | 2003-07-14 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of docking domains in modular polyketide synthases.
Chem.Biol., 10, 2003
|
|
6VJP
 
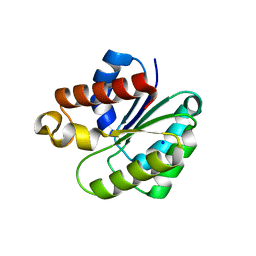 | | Structure of Staphylococcus aureus peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acetyltransferase, SODIUM ION | | Authors: | Jones, C.J, Sychantha, D, Howell, P.L, Clarke, A.J. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Structural basis for theO-acetyltransferase function of the extracytoplasmic domain of OatA fromStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
1GLB
 
 | | STRUCTURE OF THE REGULATORY COMPLEX OF ESCHERICHIA COLI IIIGLC WITH GLYCEROL KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUCOSE-SPECIFIC PROTEIN IIIGlc, GLYCEROL, ... | | Authors: | Hurley, J.H, Worthylake, D, Faber, H.R, Meadow, N.D, Roseman, S, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1992-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase.
Science, 259, 1993
|
|
6D9K
 
 | | Ternary RsAgo Complex with Guide RNA and Target DNA Containing A-G Non-canonical Pair | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, DNA (5'-D(P*TP*CP*GP*TP*CP*AP*CP*CP*TP*GP*GP*GP*CP*AP*GP*TP*AP*AP*C)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
3NJC
 
 | | Crystal structure of the yslB protein from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR460. | | Descriptor: | YSLB protein | | Authors: | Vorobiev, S, Abashidze, M, Seetharaman, J, Wang, D, Cunningham, K, Ma, L.-C, Owens, L, Fang, Y, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Crystal structure of the yslB protein from Bacillus subtilis.
To be Published
|
|
