8U45
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | Alkaline protease 1, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-09-08 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
6Y98
 
 | | Crystal Structure of subtype-switched Epithelial Adhesin 9 to 1 A domain (Epa9-CBL2Epa1) from Candida glabrata in complex with beta-lactose | | Descriptor: | CALCIUM ION, PA14 domain-containing protein, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hoffmann, D, Diderrich, R, Kock, M, Friederichs, S, Reithofer, V, Essen, L.-O, Moesch, H.-U. | | Deposit date: | 2020-03-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
J.Biol.Chem., 295, 2020
|
|
1U7P
 
 | | X-ray Crystal Structure of the Hypothetical Phosphotyrosine Phosphatase MDP-1 of the Haloacid Dehalogenase Superfamily | | Descriptor: | MAGNESIUM ION, TUNGSTATE(VI)ION, magnesium-dependent phosphatase-1 | | Authors: | Peisach, E, Selengut, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystal Structure of the Hypothetical Phosphotyrosine
Phosphatase MDP-1 of the Haloacid Dehalogenase Superfamily
Biochemistry, 43, 2004
|
|
5IS0
 
 | | Structure of TRPV1 in complex with capsazepine, determined in lipid nanodisc | | Descriptor: | Transient receptor potential cation channel subfamily V member 1, capsazepine | | Authors: | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2016-03-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
5II6
 
 | | Crystal structure of the ZP-N1 domain of mouse sperm receptor ZP2 at 0.95 A resolution | | Descriptor: | Zona pellucida sperm-binding protein 2 | | Authors: | Dioguardi, E, Han, L, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
6YS3
 
 | | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Kudlinzki, D, Hodirnau, V.V, Frangakis, A, Schwalbe, H. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structure of the 50S ribosomal subunit at 2.58 Angstroms with modeled GBC SecM peptide
Nat Commun, 2020
|
|
7N1E
 
 | | SARS-CoV-2 RLQ peptide-specific TCR pRLQ3 binds to RLQ-HLA-A2 | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, A-2 alpha chain, ... | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
7N1C
 
 | | SARS-CoV-2 RLQ peptide-specific TCR pRLQ3 | | Descriptor: | pRLQ3 T cell receptor alpha chain, pRLQ3 T cell receptor beta chain | | Authors: | Wu, D, Mariuzza, R.A. | | Deposit date: | 2021-05-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Nat Commun, 13, 2022
|
|
6Y5R
 
 | | Structure of Human Potassium Chloride Transporter KCC3 S45D/T940D/T997D in NaCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, B, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure of Human Potassium Chloride Transporter KCC3 in NaCl
To be published
|
|
5IM9
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F1 | | Descriptor: | Bacterioferritin comigratory protein, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-05 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IRE
 
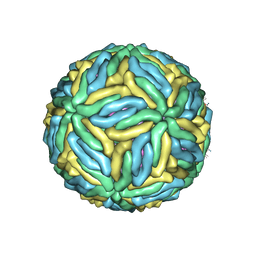 | | The cryo-EM structure of Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sirohi, D, Chen, Z, Sun, L, Klose, T, Pierson, T, Rossmann, M, Kuhn, R. | | Deposit date: | 2016-03-13 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom resolution cryo-EM structure of Zika virus.
Science, 352, 2016
|
|
5IRX
 
 | | Structure of TRPV1 in complex with DkTx and RTX, determined in lipid nanodisc | | Descriptor: | (2S)-2-(acetyloxy)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}propyl pentanoate, (2S)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, (4R,7S)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(pentanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphatetradecan-1-aminium, ... | | Authors: | Gao, Y, Cao, E, Julius, D, Cheng, Y. | | Deposit date: | 2016-03-14 | | Release date: | 2016-05-25 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Nature, 534, 2016
|
|
5IMZ
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F7 | | Descriptor: | Bacterioferritin comigratory protein, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
2UX8
 
 | | Crystal Structure of Sphingomonas elodea ATCC 31461 Glucose-1- phosphate uridylyltransferase in Complex with glucose-1-phosphate. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE URIDYLYLTRANSFERASE | | Authors: | Aragao, D, Fialho, A.M, Marques, A.R, Frazao, C, Sa-Correia, I, Mitchell, E.P. | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Complex of Sphingomonas Elodea Atcc 31461 Glucose-1-Phosphate Uridylyltransferase with Glucose-1-Phosphate Reveals a Novel Quaternary Structure, Unique Among Nucleoside Diphosphate-Sugar Pyrophosphorylase Members.
J.Bacteriol., 189, 2007
|
|
5IO2
 
 | | Xanthomonas campestris Peroxiredoxin Q - C48S mutant | | Descriptor: | Bacterioferritin comigratory protein, PHOSPHATE ION, SODIUM ION | | Authors: | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
2UYT
 
 | |
6YEZ
 
 | | Plant PSI-ferredoxin-plastocyanin supercomplex | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Caspy, I, Nelson, N, Shkolnisky, Y, Klaiman, D, Sheinker, A. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-30 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structure of a triple complex of plant photosystem I with ferredoxin and plastocyanin.
Nat.Plants, 6, 2020
|
|
1DXY
 
 | | STRUCTURE OF D-2-HYDROXYISOCAPROATE DEHYDROGENASE | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, D-2-HYDROXYISOCAPROATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dengler, U, Niefind, K, Kiess, M, Schomburg, D. | | Deposit date: | 1996-08-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a ternary complex of D-2-hydroxyisocaproate dehydrogenase from Lactobacillus casei, NAD+ and 2-oxoisocaproate at 1.9 A resolution.
J.Mol.Biol., 267, 1997
|
|
6YKZ
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_234 | | Descriptor: | SULFATE ION, YTHDC1, ~{N}-methyl-1,4,5,6-tetrahydrocyclopenta[c]pyrazole-3-carboxamide | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
1E0Z
 
 | | [2Fe-2S]-Ferredoxin from Halobacterium salinarum | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Schweimer, K, Marg, B, Oesterhelt, D, Roesch, P, Sticht, H. | | Deposit date: | 2000-04-11 | | Release date: | 2001-04-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | A Two-Alpha-Helix Extra Domain Mediates the Halophilic Character of a Plant-Type Ferredoxin from Halophilic Archaea.
Biochemistry, 44, 2005
|
|
6YKE
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_038 | | Descriptor: | (2~{R})-2-(3-fluorophenyl)-5,5-dimethyl-morpholine, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
6YL0
 
 | | Crystal structure of YTHDC1 with compound T_96 | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, YTHDC1, ... | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of YTHDC1 with compound T_96
To Be Published
|
|
8V1P
 
 | | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092 | | Descriptor: | Glucose-induced degradation protein 4 homolog, N,N~2~-bis[(4-methoxyphenyl)methyl]glycinamide | | Authors: | Dong, C, Dong, A, Calabrese, M, Wang, F, Owen, D, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-21 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092
To be published
|
|
6YL9
 
 | | Crystal structure of YTHDC1 with compound DHU_DC1_085 | | Descriptor: | 3-[(2~{R},5~{S})-2-(2,5-dimethylphenyl)-5-methyl-morpholin-4-yl]propane-1-sulfonamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Wiedmer, L, Caflisch, A. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of ligands of the m6A-RNA reader YTHDC1
Eur J Med Chem Rep, 5, 2022
|
|
5IIB
 
 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.64 A resolution (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Egg-lysin, ... | | Authors: | Raj, I, Sadat Al-Hosseini, H, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
