2Y2A
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph I | | Descriptor: | ACETATE ION, AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3UI7
 
 | | Discovery of orally active pyrazoloquinoline as a potent PDE10 inhibitor for the management of schizophrenia | | Descriptor: | 6-methoxy-3,8-dimethyl-4-(morpholin-4-ylmethyl)-1H-pyrazolo[3,4-b]quinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Yang, S, Smotryski, J, Mcelroy, W, Ho, G, Tulshian, D, Greenlee, W.J, Hodgson, R, Xiao, L, Hruza, A. | | Deposit date: | 2011-11-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of orally active pyrazoloquinolines as potent PDE10 inhibitors for the management of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
2G38
 
 | | A PE/PPE Protein Complex from Mycobacterium tuberculosis | | Descriptor: | MANGANESE (II) ION, PE FAMILY PROTEIN, PPE FAMILY PROTEIN | | Authors: | Strong, M, Sawaya, M.R, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4V0C
 
 | |
4V02
 
 | | MinC:MinD cell division protein complex, Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PROBABLE SEPTUM SITE-DETERMINING PROTEIN MINC, ... | | Authors: | Ghosal, D, Lowe, J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mincd Cell Division Proteins Form Alternating Copolymeric Cytomotive Filaments.
Nat.Commun., 5, 2014
|
|
4UXZ
 
 | | Structure of delta7-DgkA-syn in 7.9 MAG to 2.18 angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, (2S)-2,3-dihydroxypropyl (7Z)-hexadec-7-enoate, ACETATE ION, ... | | Authors: | Li, D, Howe, N, Caffrey, M. | | Deposit date: | 2014-08-27 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Ternary Structure Reveals Mechanism of a Membrane Diacylglycerol Kinase.
Nat.Commun., 6, 2015
|
|
4WAG
 
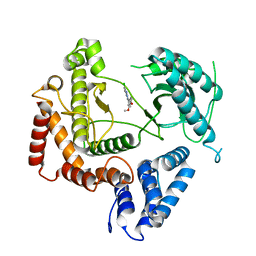 | | Phosphatidylinositol 4-kinase III beta crystallized with MI103 inhibitor | | Descriptor: | 6-chloro-3-(3,4-dimethoxyphenyl)-2-methylimidazo[1,2-b]pyridazin-8-amine, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.407 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
4WAE
 
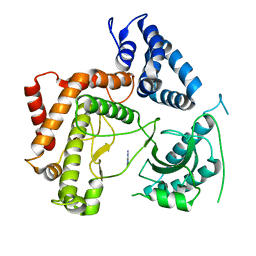 | | Phosphatidylinositol 4-kinase III beta crystallized with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.318 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
4WAB
 
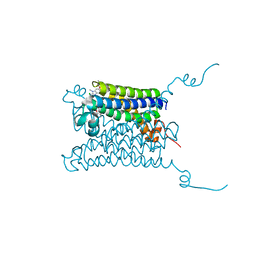 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4FN5
 
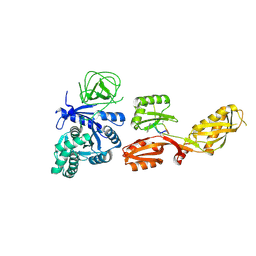 | |
7XRP
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5G2 nanobody, Spike protein S1 | | Authors: | Liu, L, Sun, H, Jiang, Y, Liu, X, Zhao, D, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2022-05-11 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode.
J Nanobiotechnology, 20, 2022
|
|
8SH4
 
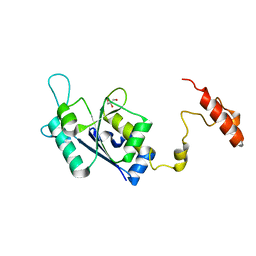 | | Crystal structure of the tRNA (m1G37) methyltransferase apoenzyme from Anaplasma phagocytophilum | | Descriptor: | GLYCEROL, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jannotta, C, Edele, D, Levanti, D, Carson, M, Prucha, G, Caesar, J, Picchiello, C, Collins, K, Garland, E, Handley-Pendleton, J, Hernandez, V, Leffler, S, Williams, D, Stojanoff, V, Perez, A, Halloran, J, Bolen, R. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the m1G37 tRNA methyltransferase apoenzyme from Anaplasma phagocytophilum
To Be Published
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
1VTD
 
 | | UNUSUAL HELICAL PACKING IN CRYSTALS OF DNA BEARING A MUTATION HOT SPOT | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*GP*CP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*GP*CP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Timsit, Y, Westhof, E, Fuchs, R.P.P, Moras, D. | | Deposit date: | 1996-12-12 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unusual helical packing in crystals of DNA bearing a mutation hot spot.
Nature, 341, 1989
|
|
5K7R
 
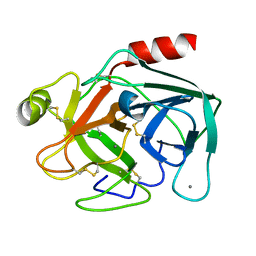 | | MicroED structure of trypsin at 1.7 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
4FBY
 
 | | fs X-ray diffraction of Photosystem II | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Hellmich, J, Tran, R, Hattne, J, Laksmono, H, Gloeckner, C, Echols, N, Sierra, R.G, Sellberg, J, Lassalle-Kaiser, B, Gildea, R.J, Glatzel, P, Grosse-Kunstleve, R.W, Latimer, M.J, Mcqueen, T.A, Difiore, D, Fry, A.R, Messerschmidt, M.M, Miahnahri, A, Schafer, D.W, Seibert, M.M, Sokaras, D, Weng, T.-C, Zwart, P.H, White, W.E, Adams, P.D, Bogan, M.J, Boutet, S, Williams, G.J, Messinger, J, Sauter, N.K, Zouni, A, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (6.56 Å) | | Cite: | Room temperature femtosecond X-ray diffraction of photosystem II microcrystals.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5K7Q
 
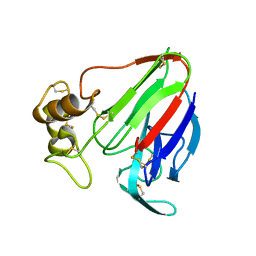 | | MicroED structure of thaumatin at 2.5 A resolution | | Descriptor: | Thaumatin-1 | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7T
 
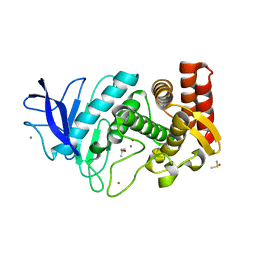 | | MicroED structure of thermolysin at 2.5 A resolution | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
6K4D
 
 | | Ancestral luciferase AncLamp in complex with ATP and D-luciferin | | Descriptor: | (4S)-2-(6-hydroxy-1,3-benzothiazol-2-yl)-4,5-dihydro-1,3-thiazole-4-carboxylic acid, Ancestral luciferase AncLamp, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] (4S)-2-(6-oxidanyl-1,3-benzothiazol-2-yl)-4,5-dihydro-1,3-thiazole-4-carboxylate | | Authors: | Oba, Y, Konishi, K, Yano, D, Kato, D, Shirai, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Resurrecting the ancient glow of the fireflies.
Sci Adv, 6, 2020
|
|
6K4C
 
 | | Ancestral luciferase AncLamp in complex with DLSA | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, Ancestral luciferase AncLamp, MAGNESIUM ION | | Authors: | Oba, Y, Konishi, K, Yano, D, Kato, D, Shirai, T. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resurrecting the ancient glow of the fireflies.
Sci Adv, 6, 2020
|
|
8R7G
 
 | | Crystal structure of the kinase domain of ACVR1 (ALK2) with M4K2234 | | Descriptor: | 2-fluoranyl-6-methoxy-4-[4-methyl-5-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pyridin-3-yl]benzamide, Activin receptor type I | | Authors: | Williams, E.P, Cros, J, Ensan, D, Smil, D, Edwards, A.M, O'Meara, J.A, Fernandez-Cid, A, Isaac, M.B, Al-awar, R, Bullock, A.N. | | Deposit date: | 2023-11-24 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Targeting ALK2: An Open Science Approach to Developing Therapeutics for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
3UYC
 
 | | Designed protein KE59 R8_2/7A | | Descriptor: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | Authors: | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-12-06 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5K7O
 
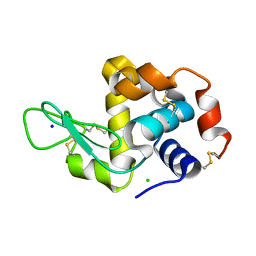 | | MicroED structure of lysozyme at 1.8 A resolution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.8 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
