6I83
 
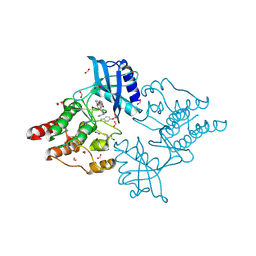 | | Crystal structure of phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018366 | | Descriptor: | 4-[5-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl]benzamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
8CPD
 
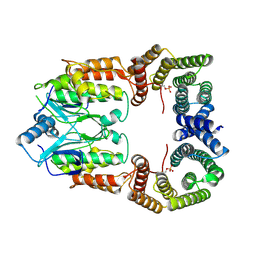 | | Cryo-EM structure of CRaf dimer with 14:3:3 | | Descriptor: | 14-3-3 protein zeta isoform X1, RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Dedden, D, Graedler, U, Schwarz, D, Thomsen, M, Leuthner, B, Schneider, E, Nitsche, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
6IAB
 
 | |
6RJ1
 
 | |
1FEP
 
 | | FERRIC ENTEROBACTIN RECEPTOR | | Descriptor: | FERRIC ENTEROBACTIN RECEPTOR | | Authors: | Buchanan, S.K, Smith, B.S, Ventatramani, L, Xia, D, Esser, L, Palnitkar, M, Chakraborty, R, Van Der Helm, D, Deisenhofer, J. | | Deposit date: | 1998-11-24 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the outer membrane active transporter FepA from Escherichia coli.
Nat.Struct.Biol., 6, 1999
|
|
2H3X
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes Faecalis (Form 3) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
3AAT
 
 | |
1CF4
 
 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
5NM4
 
 | | A2A Adenosine receptor room-temperature structure determined by serial femtosecond crystallography | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
6RHN
 
 | | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN (HINT) FROM RABBIT WITHOUT NUCLEOTIDE | | Descriptor: | HISTIDINE TRIAD NUCLEOTIDE-BINDING PROTEIN | | Authors: | Brenner, C, Garrison, P, Gilmour, J, Peisach, D, Ringe, D, Petsko, G.A, Lowenstein, J.M. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of HINT demonstrate that histidine triad proteins are GalT-related nucleotide-binding proteins.
Nat.Struct.Biol., 4, 1997
|
|
5NM2
 
 | | A2A Adenosine receptor cryo structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Weinert, T, Cheng, R, James, D, Gashi, D, Nogly, P, Jaeger, K, Dore, A.S, Geng, T, Cooke, R, Hennig, M, Standfuss, J. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons.
Nat Commun, 8, 2017
|
|
3MES
 
 | | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Choline kinase, DECAMETHONIUM ION, ... | | Authors: | Qiu, W, Wernimont, A, Hills, T, Lew, J, Artz, J.D, Xiao, T, Allali-Hassani, A, Vedadi, M, Kozieradzki, I, Cossar, D, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Hui, R, Ma, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of choline kinase from Cryptosporidium parvum Iowa II, cgd3_2030
TO BE PUBLISHED
|
|
1C4B
 
 | | A BETA-HAIRPIN MIMIC FROM FCERI-ALPHA-CYCLO(RD-262) | | Descriptor: | PROTEIN (CYCLO(RD-262)) | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Sutton, B.J, Cowburn, D. | | Deposit date: | 1999-08-02 | | Release date: | 1999-08-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of FceRI Alpha-Chain Mimics: A Beta-Hairpin Peptide and Its Retroenantiomer
J.Am.Chem.Soc., 119, 1997
|
|
2IUB
 
 | | Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. | | Descriptor: | CHLORIDE ION, DIVALENT CATION TRANSPORT-RELATED PROTEIN, MAGNESIUM ION | | Authors: | Eshaghi, S, Niegowski, D, Kohl, A, Martinez Molina, D, Lesley, S.A, Nordlund, P. | | Deposit date: | 2006-06-01 | | Release date: | 2006-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of a Divalent Metal Ion Transporter Cora at 2.9 Angstrom Resolution.
Science, 313, 2006
|
|
2J6C
 
 | | crystal structure of AFV3-109, a highly conserved protein from crenarchaeal viruses | | Descriptor: | AFV3-109, GLYCEROL | | Authors: | Keller, J, Leulliot, N, Cambillau, C, Campanacci, V, Porciero, S, Prangishvili, D, Cortez, D, Quevillon-Cheruel, S, Van Tilbeurgh, H. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Afv3-109, a Highly Conserved Protein from Crenarchaeal Viruses.
Virol J., 4, 2007
|
|
2JNA
 
 | | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109 | | Descriptor: | Putative secreted protein | | Authors: | Eletsky, A, Parish, D, Liu, G, Sukumaran, D, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-31 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109
To be Published
|
|
5AAZ
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | OPTINEURIN, ZINC ION | | Authors: | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-31 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
2ERL
 
 | | PHEROMONE ER-1 FROM | | Descriptor: | ETHANOL, MATING PHEROMONE ER-1 | | Authors: | Anderson, D.H, Weiss, M.S, Eisenberg, D. | | Deposit date: | 1995-12-20 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A challenging case for protein crystal structure determination: the mating pheromone Er-1 from Euplotes raikovi.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2EXI
 
 | | Structure of the family43 beta-Xylosidase D15G mutant from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
4CFH
 
 | | Structure of an active form of mammalian AMPK | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Sanders, M.J, Underwood, E, Heath, R, Mayer, F, Carmena, D, Jing, C, Walker, P.A, Eccleston, J.F, Haire, L.F, Saiu, P, Howell, S.A, Aasland, R, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2013-11-18 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structure of Mammalian Ampk and its Regulation by Adp
Nature, 472, 2011
|
|
2EZ1
 
 | | Holo tyrosine phenol-lyase from Citrobacter freundii at pH 8.0 | | Descriptor: | POTASSIUM ION, Tyrosine phenol-lyase | | Authors: | Milic, D, Matkovic-Calogovic, D, Demidkina, T.V, Antson, A.A. | | Deposit date: | 2005-11-10 | | Release date: | 2006-07-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of apo- and holo-tyrosine phenol-lyase reveal a catalytically critical closed conformation and suggest a mechanism for activation by K+ ions
Biochemistry, 45, 2006
|
|
2F8U
 
 | | G-quadruplex structure formed in human Bcl-2 promoter, hybrid form | | Descriptor: | 5'-D(*GP*GP*GP*CP*GP*CP*GP*GP*GP*AP*GP*GP*AP*AP*TP*TP*GP*GP*GP*CP*GP*GP*G)-3' | | Authors: | Dai, J, Chen, D, Carver, M, Yang, D. | | Deposit date: | 2005-12-03 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region.
Nucleic Acids Res., 34, 2006
|
|
5AAY
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | NF-KAPPA-B ESSENTIAL MODULATOR, ZINC ION | | Authors: | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-31 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
3NMQ
 
 | | Hsp90b N-terminal domain in complex with EC44, a pyrrolo-pyrimidine methoxypyridine inhibitor | | Descriptor: | 5-{2-amino-4-chloro-7-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-7H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-methylpent-4-yn-2 -ol, Heat shock protein HSP 90-beta | | Authors: | Arndt, J.W, Yun, T.J, Harning, E.K, Giza, K, Rabah, D, Li, P, Luchetti, D, Shi, J, Manning, A, Kehry, M.R. | | Deposit date: | 2010-06-22 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | EC144, a Synthetic Inhibitor of Heat Shock Protein 90, Blocks Innate and Adaptive Immune Responses in Models of Inflammation and Autoimmunity.
J.Immunol., 186, 2011
|
|
2JY5
 
 | | NMR structure of Ubiquilin 1 UBA domain | | Descriptor: | Ubiquilin-1 | | Authors: | Zhang, D, Raasi, S, Fushman, D. | | Deposit date: | 2007-12-06 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains
J.Mol.Biol., 377, 2008
|
|
