6SZP
 
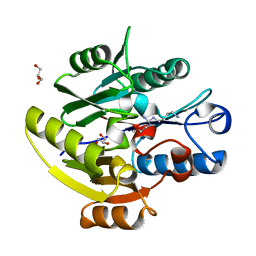 | | High resolution crystal structure of human DDAH-1 in complex with N-(4-Aminobutyl)-N'-(2-Methoxyethyl)guanidine | | Descriptor: | (1~{S})-~{N}'-(4-azanylbutyl)-~{N}"-(2-methoxyethyl)methanetriamine, GLYCEROL, N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Hennig, S, Vetter, I.R, Schade, D. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery ofN-(4-Aminobutyl)-N'-(2-methoxyethyl)guanidine as the First Selective, Nonamino Acid, Catalytic Site Inhibitor of Human Dimethylarginine Dimethylaminohydrolase-1 (hDDAH-1).
J.Med.Chem., 63, 2020
|
|
3K2Y
 
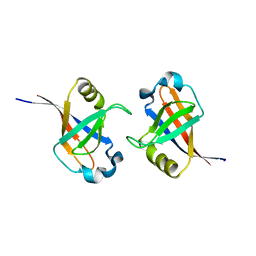 | | Crystal structure of protein lp_0118 from Lactobacillus plantarum,northeast structural genomics consortium target LpR91B | | Descriptor: | uncharacterized protein lp_0118 | | Authors: | Seetharaman, J, Lew, S, Vorobiev, S.M, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of protein lp_0118 from Lactobacillus plantarum,northeast structural genomics consortium target LpR91B
To be Published
|
|
6SYZ
 
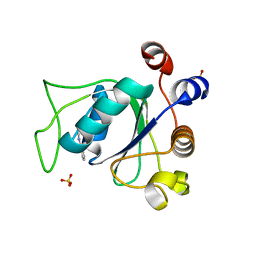 | | Crystal structure of YTHDC1 with fragment 1 (DHU_DC1_141) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methylthieno[3,2-d]pyrimidin-4-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T01
 
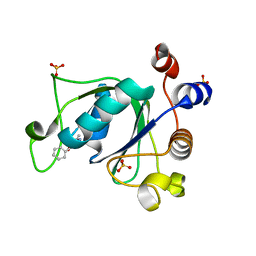 | | Crystal structure of YTHDC1 with fragment 13 (DHU_DC1_153) | | Descriptor: | 6-methyl-5-nitro-4-phenyl-1~{H}-pyrimidin-2-one, SULFATE ION, YTH domain-containing protein 1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
6T0A
 
 | | Crystal structure of YTHDC1 with fragment 25 (PSI_DC1_005) | | Descriptor: | (~{R})-azanyl(pyridin-3-yl)methanethiol, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3K74
 
 | | Disruption of protein dynamics by an allosteric effector antibody | | Descriptor: | Dihydrofolate reductase, Nanobody | | Authors: | Oyen, D, Srinivasan, V, Steyaert, J, Barlow, J. | | Deposit date: | 2009-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Constraining enzyme conformational change by an antibody leads to hyperbolic inhibition.
J.Mol.Biol., 407, 2011
|
|
6FKK
 
 | | Drosophila Semaphorin 1b, extracellular domains 1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MIP07328p, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rozbesky, D, Harlos, K, Jones, E.Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6SL5
 
 | | Dunaliella Photosystem I Supercomplex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nelson, N, Caspy, I, Malavath, T, Klaiman, D, Shkolinsky, Y. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure and energy transfer pathways of the Dunaliella Salina photosystem I supercomplex.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
6FDB
 
 | |
3KA5
 
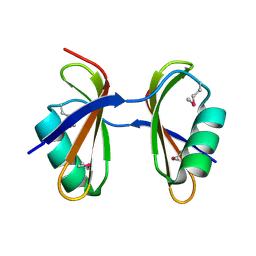 | | Crystal structure of Ribosome-associated protein Y (PSrp-1) from Clostridium acetobutylicum. Northeast Structural Genomics Consortium target id CaR123A | | Descriptor: | Ribosome-associated protein Y (PSrp-1) | | Authors: | Seetharaman, J, Neely, H, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-18 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Ribosome-associated protein Y (PSrp-1) from Clostridium acetobutylicum
To be Published
|
|
3KX3
 
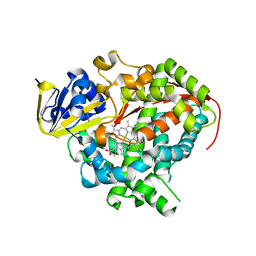 | | Crystal structure of Bacillus megaterium BM3 heme domain mutant L86E | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, N-PALMITOYLGLYCINE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Girvan, H.M, Levy, C.W, Leys, D, Munro, A.W. | | Deposit date: | 2009-12-02 | | Release date: | 2010-05-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Glutamate-haem ester bond formation is disfavoured in flavocytochrome P450 BM3: characterization of glutamate substitution mutants at the haem site of P450 BM3.
Biochem.J., 427, 2010
|
|
6SS1
 
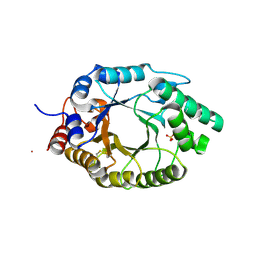 | | Kemp Eliminase HG3.17 mutant Q50A, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50A, E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6FKP
 
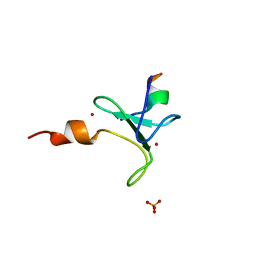 | | Crystal structure of BAZ2A PHD zinc finger in complex with H3 10-mer AA mutant peptide | | Descriptor: | ALA-ARG-THR-ALA-ALA-THR-ALA-ARG, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
6FH6
 
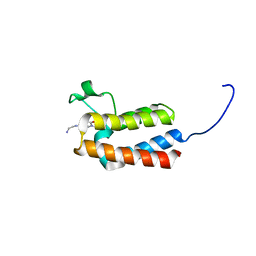 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methylpyridinone compound 1 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-(2-azanylethyl)-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-12 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6STB
 
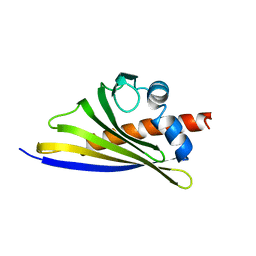 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, Q64W mutant | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
6SU6
 
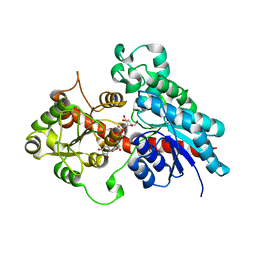 | | Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and UDP-glucose | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Fredslund, F, Teze, D, Svensson, B, Adams, P.D, Welner, D.H. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | O-/N-/S-Specificity in Glycosyltransferase Catalysis: From Mechanistic Understanding to Engineering
Acs Catalysis, 11, 2021
|
|
3KVE
 
 | | Structure of native L-amino acid oxidase from Vipera ammodytes ammodytes: stabilization of the quaternary structure by divalent ions and structural changes in the dynamic active site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase, ... | | Authors: | Gergiova, D, Murakami, M.T, Perbandt, M, Arni, R.K, Betzel, C. | | Deposit date: | 2009-11-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure of native L-amino acid oxidase from Vipera ammodytes ammodytes: stabilization of the quaternary structure by divalent ions and structural changes in the dynamic active site
To be Published
|
|
6SS3
 
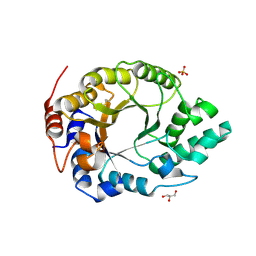 | | Kemp Eliminase HG3.17 mutant Q50K, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50K, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6SU3
 
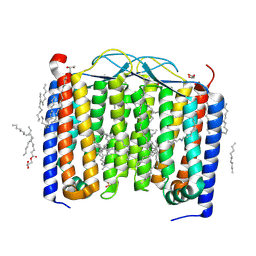 | | Crystal structure of the 48C12 heliorhodopsin in the violet form at pH 8.8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 48C12 heliorhodopsin, EICOSANE, ... | | Authors: | Kovalev, K, Volkov, D, Astashkin, R, Alekseev, A, Gushchin, I, Gordeliy, V. | | Deposit date: | 2019-09-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural insights into the heliorhodopsin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3KHD
 
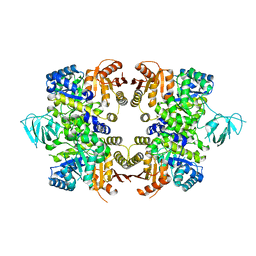 | | Crystal Structure of PFF1300w. | | Descriptor: | Pyruvate kinase | | Authors: | Wernimont, A.K, Hutchinson, A, Hassanali, A, Mackenzie, F, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Pizarro, J.C, Bakszt, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of PFF1300w.
To be Published
|
|
6SYK
 
 | | Guanine-rich oligonucleotide with 5'- and 3'-GC ends form G-quadruplex with A(GGGG)A hexad, GCGC- and G-quartets and two symmetric GG and AA base pair | | Descriptor: | GCnCG | | Authors: | Pavc, D, Wang, B, Spindler, L, Drevensek-Olenik, I, Plavec, J, Sket, P. | | Deposit date: | 2019-09-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | GC ends control topology of DNA G-quadruplexes and their cation-dependent assembly.
Nucleic Acids Res., 48, 2020
|
|
6SUG
 
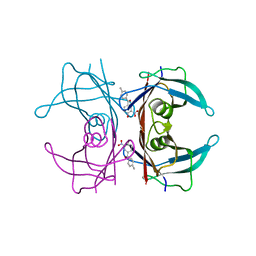 | | Crystal structure of transthyretin in complex with 3-deoxytolcapone, a tolcapone analogue | | Descriptor: | 3-deoxytolcapone, Transthyretin | | Authors: | Loconte, V, Cianci, M, Menozzi, I, Sbravati, D, Sansone, F, Casnati, A, Berni, R. | | Deposit date: | 2019-09-14 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Interactions of tolcapone analogues as stabilizers of the amyloidogenic protein transthyretin.
Bioorg.Chem., 103, 2020
|
|
6FCE
 
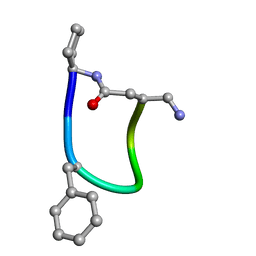 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | Descriptor: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-23 | | Method: | SOLUTION NMR | | Cite: | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
3KJS
 
 | | Crystal Structure of T. cruzi DHFR-TS with 3 high affinity DHFR inhibitors: DQ1 inhibitor complex | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase-thymidylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2009-11-03 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and characterization of potent inhibitors of Trypanosoma cruzi dihydrofolate reductase.
Bioorg.Med.Chem., 18, 2010
|
|
6SRY
 
 | | Kemp Eliminase HG3.17 mutant Q50S, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50S, E47N,N300D, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
