4RL6
 
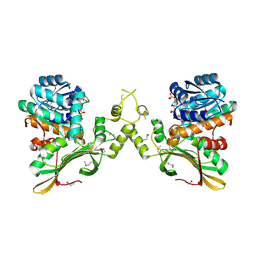 | | Crystal Structure of the Q04L03_STRP2 protein from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR105 | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase | | Authors: | Vorobiev, S, Neely, H.M, Odukwe, C.D, Seetharaman, J, Mao, L, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure of the Q04L03_STRP2 protein from Streptococcus pneumoniae.
To be Published
|
|
5W4K
 
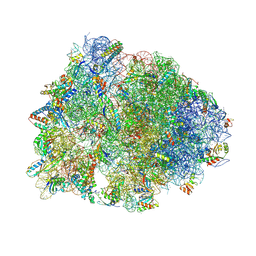 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Klebsazolicin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Metelev, M, Osterman, I.A, Ghilarov, D, Khabibullina, N.F, Yakimov, A, Shabalin, K, Utkina, I, Travin, D.Y, Komarova, E.S, Serebryakova, M, Artamonova, T, Khodorkovskii, M, Konevega, A.L, Sergiev, P.V, Severinov, K, Polikanov, Y.S. | | Deposit date: | 2017-06-12 | | Release date: | 2017-08-30 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.
Nat. Chem. Biol., 13, 2017
|
|
4RKS
 
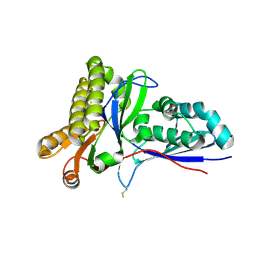 | | Crystal Structure of Mevalonate-3-Kinase from Thermoplasma acidophilum (Mevalonate Bound) | | Descriptor: | (R)-MEVALONATE, ACETATE ION, GLYCEROL, ... | | Authors: | Vinokur, J.M, Cascio, D, Sawaya, M.R, Bowie, J.U. | | Deposit date: | 2014-10-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of mevalonate-3-kinase provides insight into the mechanisms of isoprenoid pathway decarboxylases.
Protein Sci., 24, 2015
|
|
4RS8
 
 | |
2HSP
 
 | | SOLUTION STRUCTURE OF THE SH3 DOMAIN OF PHOSPHOLIPASE CGAMMA | | Descriptor: | PHOSPHOLIPASE C-GAMMA (SH3 DOMAIN) | | Authors: | Kohda, D, Hatanaka, H, Odaka, M, Inagaki, F. | | Deposit date: | 1994-06-13 | | Release date: | 1994-08-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of phospholipase C-gamma.
Cell(Cambridge,Mass.), 72, 1993
|
|
4FVF
 
 | |
2HLP
 
 | | CRYSTAL STRUCTURE OF THE E267R MUTANT OF A HALOPHILIC MALATE DEHYDROGENASE IN THE APO FORM | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, SODIUM ION | | Authors: | Richard, S.B, Madern, D, Garcin, E, Zaccai, G. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Halophilic adaptation: novel solvent protein interactions observed in the 2.9 and 2.6 A resolution structures of the wild type and a mutant of malate dehydrogenase from Haloarcula marismortui.
Biochemistry, 39, 2000
|
|
2HOX
 
 | | alliinase from allium sativum (garlic) | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shimon, L.J.W, Rabinkov, A, Wilcheck, M, Mirelman, D, Frolow, F. | | Deposit date: | 2006-07-17 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two Structures of Alliinase from Alliium sativum L.: Apo Form and Ternary Complex with Aminoacrylate Reaction Intermediate Covalently Bound to the PLP Cofactor.
J.Mol.Biol., 366, 2007
|
|
4FVG
 
 | |
4JDD
 
 | |
4ONG
 
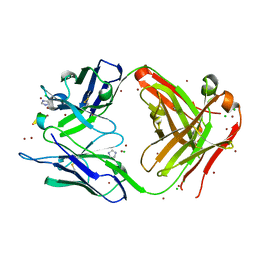 | | Fab fragment of 3D6 in complex with amyloid beta 1-40 | | Descriptor: | 3D6 FAB ANTIBODY HEAVY CHAIN, 3D6 FAB ANTIBODY LIGHT CHAIN, Amyloid beta A4 protein, ... | | Authors: | Feinberg, H, Saldanha, J.W, Diep, L, Goel, A, Widom, A, Veldman, G.M, Weis, W.I, Schenk, D, Basi, G.S. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure reveals conservation of amyloid-beta conformation recognized by 3D6 following humanization to bapineuzumab.
Alzheimers Res Ther, 6, 2014
|
|
2LBD
 
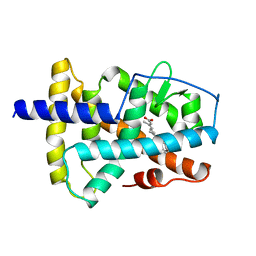 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO ALL-TRANS RETINOIC ACID | | Descriptor: | RETINOIC ACID, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Renaud, J.-P, Rochel, N, Ruff, M, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1997-08-19 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid.
Nature, 378, 1995
|
|
4I0X
 
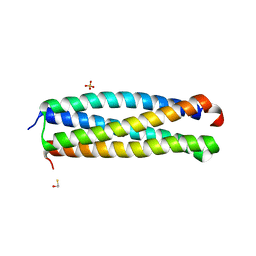 | | Crystal structure of the Mycobacterum abscessus EsxEF (Mab_3112-Mab_3113) complex | | Descriptor: | BETA-MERCAPTOETHANOL, ESAT-6-like protein MAB_3112, ESAT-6-like protein MAB_3113, ... | | Authors: | Arbing, M.A, Chan, S, Lu, J, Kuo, E, Harris, L, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-11-19 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Heterologous expression of mycobacterial Esx complexes in Escherichia coli for structural studies is facilitated by the use of maltose binding protein fusions.
Plos One, 8, 2013
|
|
3WIS
 
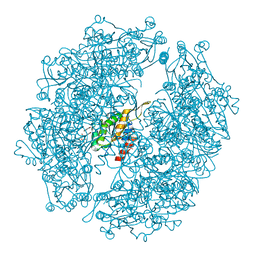 | | Crystal structure of Burkholderia xenovorans DmrB in complex with FMN: A Cubic Protein Cage for Redox Transfer | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative dihydromethanopterin reductase (AfpA), SULFATE ION | | Authors: | Bobik, T.A, Cascio, D, Jorda, J, McNamara, D.E, Bustos, C, Wang, T.C, Rasche, M.E, Yeates, T.O. | | Deposit date: | 2013-09-25 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of dihydromethanopterin reductase, a cubic protein cage for redox transfer
J.Biol.Chem., 289, 2014
|
|
4JC3
 
 | |
4JJ3
 
 | |
2JCW
 
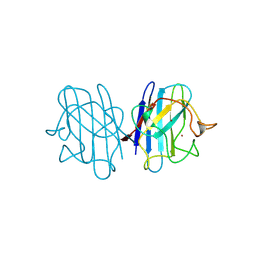 | | REDUCED BRIDGE-BROKEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (I) ION, CU/ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-21 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
2J9X
 
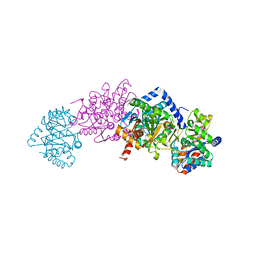 | | Tryptophan Synthase in complex with GP, alpha-D,L-glycerol-phosphate, Cs, pH6.5 - alpha aminoacrylate form - (GP)E(A-A) | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Ngo, H, Kimmich, N, Harris, R, Niks, D, Blumenstein, L, Kulik, V, Barends, T.R, Schlichting, I, Dunn, M.F. | | Deposit date: | 2006-11-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Regulation of Substrate Channeling in Tryptophan Synthase: Modulation of the L-Serine Reaction in Stage I of the Beta-Reaction by Alpha-Site Ligands.
Biochemistry, 46, 2007
|
|
4JLG
 
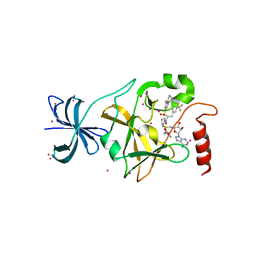 | | SETD7 in complex with inhibitor (R)-PFI-2 and S-adenosyl-methionine | | Descriptor: | 8-fluoro-N-{(2R)-1-oxo-1-(pyrrolidin-1-yl)-3-[3-(trifluoromethyl)phenyl]propan-2-yl}-1,2,3,4-tetrahydroisoquinoline-6-sulfonamide, Histone-lysine N-methyltransferase SETD7, S-ADENOSYLMETHIONINE, ... | | Authors: | Dong, A, Wu, H, Zeng, H, El Bakkouri, M, Barsyte, D, Vedadi, M, Tatlock, J, Owen, D, Bunnage, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4JRA
 
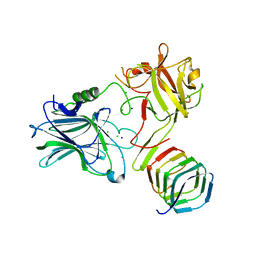 | | CRYSTAL STRUCTURE OF THE BOTULINUM NEUROTOXIN A RECEPTOR-BINDING DOMAIN IN COMPLEX WITH THE LUMINAL DOMAIN Of SV2C | | Descriptor: | Botulinum neurotoxin type A, CHLORIDE ION, SODIUM ION, ... | | Authors: | Benoit, R.M, Frey, D, Wieser, M.M, Jaussi, R, Schertler, G.F.X, Capitani, G, Kammerer, R.A. | | Deposit date: | 2013-03-21 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of synaptic vesicle protein 2C by botulinum neurotoxin A.
Nature, 505, 2014
|
|
2JAE
 
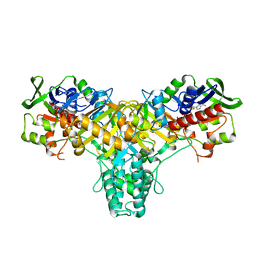 | | The structure of L-amino acid oxidase from Rhodococcus opacus in the unbound state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-AMINO ACID OXIDASE | | Authors: | Faust, A, Niefind, K, Hummel, W, Schomburg, D. | | Deposit date: | 2006-11-27 | | Release date: | 2007-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Structure of a Bacterial L-Amino Acid Oxidase from Rhodococcus Opacus Gives New Evidence for the Hydride Mechanism for Dehydrogenation.
J.Mol.Biol., 367, 2007
|
|
2K4L
 
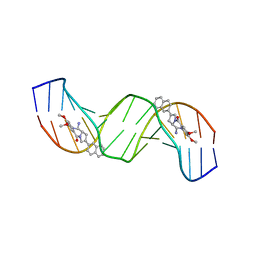 | | Solution structure of a 2:1C2-(2-naphthyl)pyrrolo[2,1-c][1,4]benzodiazepine (PBD) DNA adduct: molecular basis for unexpectedly high DNA helix stabilization. | | Descriptor: | (11aS)-7,8-dimethoxy-2-naphthalen-2-yl-1,10,11,11a-tetrahydro-5H-pyrrolo[2,1-c][1,4]benzodiazepin-5-one, 5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DAP*DAP*DAP*DGP*DAP*DTP*DT)-3' | | Authors: | Antonow, D, Barata, T, Jenkins, T.C, Parkinson, G.N, Howard, P.W, Thurston, D.E, Zloh, M. | | Deposit date: | 2008-06-13 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a 2:1 C2-(2-naphthyl) pyrrolo[2,1-c][1,4]benzodiazepine DNA adduct: molecular basis for unexpectedly high DNA helix stabilization.
Biochemistry, 47, 2008
|
|
4JJ0
 
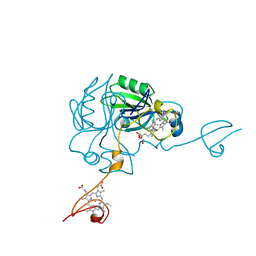 | | Crystal structure of MamP | | Descriptor: | GLYCEROL, HEME C, MamP | | Authors: | Siponen, M, Pignol, D, Arnoux, P. | | Deposit date: | 2013-03-07 | | Release date: | 2013-10-09 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into magnetochrome-mediated magnetite biomineralization.
Nature, 502, 2013
|
|
2LJ4
 
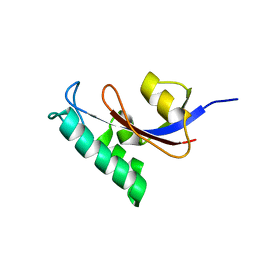 | | Solution structure of the TbPIN1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase/rotamase, putative | | Authors: | Sun, L, Lin, D, Zhao, Y. | | Deposit date: | 2011-09-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structural analysis of the single-domain parvulin TbPin1.
Plos One, 7, 2012
|
|
2L6E
 
 | | NMR Structure of the monomeric mutant C-terminal domain of HIV-1 Capsid in complex with stapled peptide Inhibitor | | Descriptor: | Capsid protein p24, NYAD-13 stapled peptide inhibitor | | Authors: | Bhattacharya, S, Zhang, H, Debnath, A.K, Cowburn, D. | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid.
J.Biol.Chem., 283, 2008
|
|
