1S9S
 
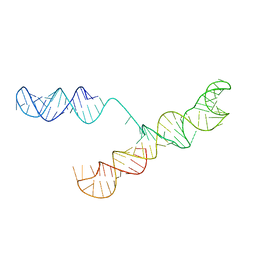 | |
6JX6
 
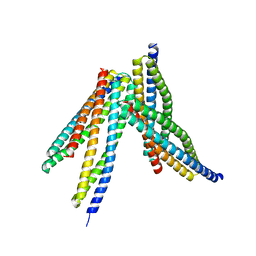 | | Tetrameric form of Smac | | Descriptor: | Diablo homolog, mitochondrial | | Authors: | Sivaraman, J, Singh, S, Ng, J, Nayak, D. | | Deposit date: | 2019-04-22 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Structural insights into a HECT-type E3 ligase AREL1 and its ubiquitination activitiesin vitro.
J.Biol.Chem., 294, 2019
|
|
1S9Z
 
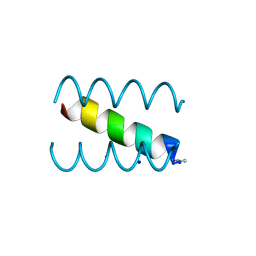 | | SYNTHETIC 17 AMINO ACID LONG PEPTIDE THAT FORMS A NATIVE-LIKE COILED-COIL AT AMBIENT TEMPERATURE AND AGGREGATES INTO AMYLOID-LIKE FIBRILS AT HIGHER TEMPERATURES. | | Descriptor: | SODIUM ION, SYNTHETIC COILED-COIL PEPTIDE, ZINC ION | | Authors: | Kammerer, R.A, Kostrewa, D, Zurdo, J, Detken, A, Garcia-Echeverria, C, Green, J.D, Muller, S.A, Meier, B.H, Winkler, F.K, Dobson, C.M, Steinmetz, M.O. | | Deposit date: | 2004-02-06 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Exploring amyloid formation by a de novo design
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3MFB
 
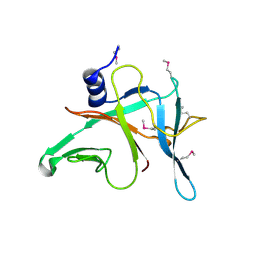 | | Crystal Structure of the S-type Pyocin domain of ECA1669 protein from Erwinia carotovora, Northeast Structural Genomics Consortium Target EwR82C | | Descriptor: | Uncharacterized protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-01 | | Release date: | 2010-04-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target EwR82C
To be Published
|
|
6K1I
 
 | | Human nucleosome core particle with gammaH2A.X variant | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2AX, ... | | Authors: | Sharma, D, De Falco, L, Davey, C.A. | | Deposit date: | 2019-05-10 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | PARP1 exhibits enhanced association and catalytic efficiency with gamma H2A.X-nucleosome.
Nat Commun, 10, 2019
|
|
1SCD
 
 | |
6K42
 
 | | cryo-EM structure of alpha2BAR-Gi1 complex | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor,Endolysin,Alpha-2B adrenergic receptor,Alpha-2B adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, D, Liu, Z, Wang, H.W, Kobilka, B.K. | | Deposit date: | 2019-05-23 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Activation of the alpha2Badrenoceptor by the sedative sympatholytic dexmedetomidine.
Nat.Chem.Biol., 16, 2020
|
|
1SF3
 
 | | Structure of the reduced form of the P94A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION, PHOSPHATE ION | | Authors: | Carrell, C.J, Sun, D, Jiang, S, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Studies of Two Mutants of Amicyanin from Paracoccus denitrificans That Stabilize the Reduced State of the Copper.
Biochemistry, 43, 2004
|
|
1SMH
 
 | | Protein kinase A variant complex with completely ordered N-terminal helix | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-OCTANOYL-N-METHYLGLUCAMINE, cAMP-Dependent Protein Kinase, ... | | Authors: | Breitenlechner, C, Engh, R.A, Huber, R, Kinzel, V, Bossemeyer, D, Gassel, M. | | Deposit date: | 2004-03-09 | | Release date: | 2004-07-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.044 Å) | | Cite: | The Typically Disordered N-Terminus of PKA Can Fold as a Helix and Project the Myristoylation Site into Solution
Biochemistry, 43, 2004
|
|
3LZW
 
 | | Crystal structure of ferredoxin-NADP+ oxidoreductase from bacillus subtilis (form I) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Komori, H, Seo, D, Sakurai, T, Higuchi, Y. | | Deposit date: | 2010-03-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity
Protein Sci., 19, 2010
|
|
3MRK
 
 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with AFP137 nonapeptide | | Descriptor: | 9-meric peptide from Alpha-fetoprotein, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Chouquet, A, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of MHC class I HLA-A2 molecule complexed with AFP137 nonapeptide
To be Published
|
|
7T9L
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
6K7F
 
 | | Crystal structure of MBPholo-Tim21 fusion protein with a 17-residue helical linker | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Mitochondrial import inner membrane translocase subunit TIM21, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bala, S, Shimada, A, Kohda, D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
4K8G
 
 | | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans mutant (V161A, R163A, K165G, L166A, Y167G, Y168A, E169G) | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Lukk, T, Wichelecki, D, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2013-04-18 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of D-Mannonate dehydratase from Novosphingobium aromaticivorans mutant (V161A, R163A, K165G, L166A, Y167G, Y168A, E169G)
To be Published
|
|
7SY2
 
 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXR
 
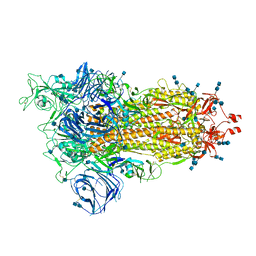 | | Cryo-EM structure of the SARS-CoV-2 D614G mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
3M3Y
 
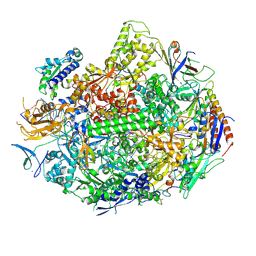 | | RNA polymerase II elongation complex C | | Descriptor: | DNA (28-MER), DNA (5'-D(*GP*TP*GP*GP*TP*TP*AP*TP*GP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Zhu, G, Huang, X, Lippard, S.J. | | Deposit date: | 2010-03-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7SY4
 
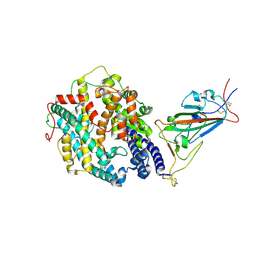 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SY6
 
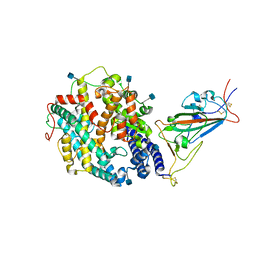 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417N mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXT
 
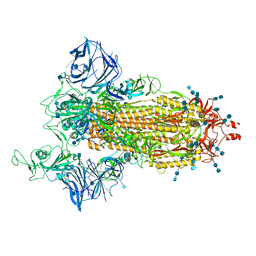 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
3M7S
 
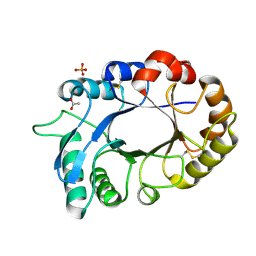 | | Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, S, Dube, D, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
7SY7
 
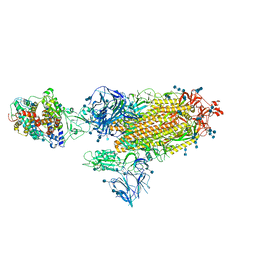 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417T mutant spike protein ectodomain bound to human ACE2 ectodomain (global refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7T9J
 
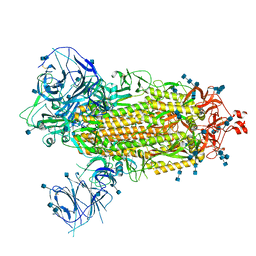 | | Cryo-EM structure of the SARS-CoV-2 Omicron spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
7SXS
 
 | | Cryo-EM structure of the SARS-CoV-2 D614G,L452R mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
1SDI
 
 | | 1.65 A structure of Escherichia coli ycfC gene product | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Hypothetical protein ycfC | | Authors: | Borek, D, Otwinowski, Z, Chen, Y, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of Escherichia coli ycfC gene product
To be Published
|
|
