1S2V
 
 | | Crystal structure of phosphoenolpyruvate mutase complexed with Mg(II) | | Descriptor: | MAGNESIUM ION, Phosphoenolpyruvate phosphomutase | | Authors: | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-01-11 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
6K3B
 
 | | Crystal structure of Lpg2147-Lpg2149 complex | | Descriptor: | Lpg2147, Uncharacterized protein | | Authors: | Mu, Y, Wang, Y, Han, Y, Li, D, Feng, Y. | | Deposit date: | 2019-05-17 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
6K41
 
 | | cryo-EM structure of alpha2BAR-GoA complex | | Descriptor: | 4-[(1~{S})-1-(2,3-dimethylphenyl)ethyl]-1~{H}-imidazole, Alpha-2A adrenergic receptor,Endolysin,Alpha-2B adrenergic receptor,Alpha-2B adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yuan, D, Liu, Z, Wang, H.W, Kobilka, B.K. | | Deposit date: | 2019-05-23 | | Release date: | 2020-04-15 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of the alpha2Badrenoceptor by the sedative sympatholytic dexmedetomidine.
Nat.Chem.Biol., 16, 2020
|
|
1SM7
 
 | | Solution structure of the recombinant pronapin precursor, BnIb. | | Descriptor: | recombinant Ib pronapin | | Authors: | Pantoja-Uceda, D, Palomares, O, Bruix, M, Villalba, M, Rodriguez, R, Rico, M, Santoro, J. | | Deposit date: | 2004-03-08 | | Release date: | 2005-02-01 | | Last modified: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties.
Biochemistry, 43, 2004
|
|
1SA9
 
 | | Crystal Structure of the RNA octamer GGCGAGCC | | Descriptor: | 5'-R(*GP*GP*CP*GP*AP*GP*CP*C)-3' | | Authors: | Jang, S.B, Baeyens, K, Jeong, M.S, SantaLucia Jr, J, Turner, D, Holbrook, S.R. | | Deposit date: | 2004-02-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structures of two RNA octamers containing tandem G.A base pairs.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SG3
 
 | | Structure of allantoicase | | Descriptor: | Allantoicase | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Sorel, I, Graille, M, Meyer, P, Liger, D, Blondeau, K, Janin, J, van Tilbeurgh, H. | | Deposit date: | 2004-02-23 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Yeast Allantoicase Reveals a Repeated Jelly Roll Motif
J.Biol.Chem., 279, 2004
|
|
4KAC
 
 | | X-Ray Structure of the complex HaloTag2 with HALTS. Northeast Structural Genomics Consortium (NESG) Target OR150. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AMMONIUM ION, Haloalkane dehalogenase, ... | | Authors: | Kuzin, A, Lew, S, Neklesa, T.K, Noblin, D, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, H, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Crews, C.M, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-22 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR150
To be Published
|
|
1SL3
 
 | | crystal structue of Thrombin in complex with a potent P1 heterocycle-Aryl based inhibitor | | Descriptor: | (2-[6-CHLORO-3-{[2,2-DIFLUORO-2-(1-OXIDOPYRIDIN-2-YL)ETHYL]AMINO}-2-OXOPYRAZIN-1(2H)-YL]-N-[5-CHLORO-2-(1H-TETRAZOL-1-YL)BENZYL]ACETAMIDE, Hirudin, thrombin | | Authors: | Young, M.B, Barrow, J.C, Glass, K.L, Lundell, G.F, Newton, C.L, Pellicore, J.M, Rittle, K.E, Selnick, H.G, Stauffer, K.J, Vacca, J.P, Williams, P.D, Bohn, D, Clayton, F.C, Cook, J.J, Krueger, J.A, Kuo, L.C, Lewis, S.D, Lucas, B.J, McMasters, D.R, Miller-Stein, C, Pietrak, B.L. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-03 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery and evaluation of potent P1 aryl heterocycle-based thrombin inhibitors
J.Med.Chem., 47, 2004
|
|
3MA2
 
 | | Complex membrane type-1 matrix metalloproteinase (MT1-MMP) with tissue inhibitor of metalloproteinase-1 (TIMP-1) | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-14, Metalloproteinase inhibitor 1, ... | | Authors: | Grossman, M, Tworowski, D, Dym, O, Lee, M.-H, Levy, Y, Sagi, I. | | Deposit date: | 2010-03-23 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Intrinsic Protein Flexibility of Endogenous Protease Inhibitor TIMP-1 Controls Its Binding Interface and Affects Its Function.
Biochemistry, 49, 2010
|
|
1SP4
 
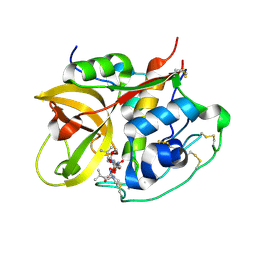 | | Crystal structure of NS-134 in complex with bovine cathepsin B: a two headed epoxysuccinyl inhibitor extends along the whole active site cleft | | Descriptor: | Cathepsin B, methyl N-[(2S)-4-{[(1S)-1-{[(2S)-2-carboxypyrrolidin-1-yl]carbonyl}-3-methylbutyl]amino}-2-hydroxy-4-oxobutanoyl]-L-leucylglycylglycinate | | Authors: | Stern, I, Schaschke, N, Moroder, L, Turk, D. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of NS-134 in complex with bovine cathepsin B: a two-headed epoxysuccinyl inhibitor extends along the entire active-site cleft.
Biochem.J., 381, 2004
|
|
1SR6
 
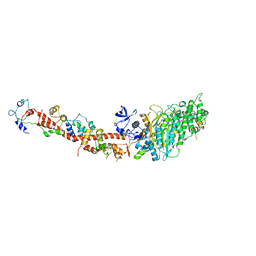 | | Structure of nucleotide-free scallop myosin S1 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Risal, D, Gourinath, S, Himmel, D.M, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6K04
 
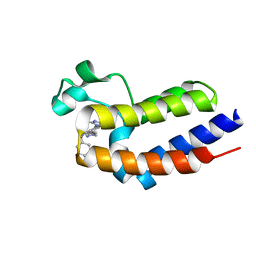 | | Crystal structure of BRD2(BD2)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
3MRE
 
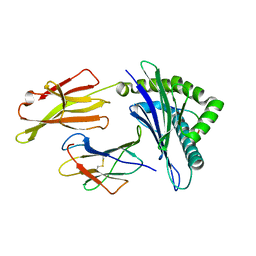 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with EBV bmlf1-280-288 nonapeptide | | Descriptor: | 9-meric peptide from mRNA export factor EB2, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
3MRL
 
 | | Crystal Structure of MHC class I HLA-A2 molecule complexed with HCV NS3-1073-1081 nonapeptide C6V variant | | Descriptor: | 9-meric peptide from Serine protease/NTPase/helicase NS3, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Reiser, J.-B, Chouquet, A, Le Gorrec, M, Debeaupuis, E, Echasserieau, K, Saulquin, X, Bonneville, M, Housset, D. | | Deposit date: | 2010-04-29 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Analysis of Relationships between Peptide/MHC Structural Features and Naive T Cell Frequency in Humans.
J.Immunol., 193, 2014
|
|
1S9P
 
 | | crystal structure of the ligand-binding domain of the estrogen-related receptor gamma in complex with diethylstilbestrol | | Descriptor: | DIETHYLSTILBESTROL, Estrogen-related receptor gamma | | Authors: | Greschik, H, Flaig, R, Renaud, J.P, Moras, D. | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis for the Deactivation of the Estrogen-related Receptor {gamma} by Diethylstilbestrol or 4-Hydroxytamoxifen and Determinants of Selectivity.
J.Biol.Chem., 279, 2004
|
|
1SCB
 
 | | ENZYME CRYSTAL STRUCTURE IN A NEAT ORGANIC SOLVENT | | Descriptor: | ACETONITRILE, CALCIUM ION, SUBTILISIN CARLSBERG | | Authors: | Fitzpatrick, P.A, Steinmetz, A.C.U, Ringe, D, Klibanov, A.M. | | Deposit date: | 1993-07-13 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme crystal structure in a neat organic solvent.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1S5M
 
 | | Xylose Isomerase in Substrate and Inhibitor Michaelis States: Atomic Resolution Studies of a Metal-Mediated Hydride Shift | | Descriptor: | MANGANESE (II) ION, SODIUM ION, Xylose isomerase, ... | | Authors: | Fenn, T.D, Ringe, D, Petsko, G.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Xylose isomerase in substrate and inhibitor michaelis States: atomic resolution studies of a metal-mediated hydride shift(,).
Biochemistry, 43, 2004
|
|
1SFH
 
 | | Reduced state of amicyanin mutant P94F | | Descriptor: | Amicyanin, COPPER (I) ION, SODIUM ION | | Authors: | Carrell, C.J, Sun, D, Jiang, S, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2004-02-19 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Studies of Two Mutants of Amicyanin from Paracoccus denitrificans That Stabilize the Reduced State of the Copper.
Biochemistry, 43, 2004
|
|
6KGZ
 
 | | bacterial cystathionine gamma-lyase MccB of Staphylococcus aureus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cystathionine gamma-lyase | | Authors: | Ha, N.-C, Lee, D. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Bacterial Cystathionine Gamma-Lyase in The Cysteine Biosynthesis Pathway of Staphylococcus aureus
Crystals, 9, 2019
|
|
1SFT
 
 | | ALANINE RACEMASE | | Descriptor: | ACETATE ION, ALANINE RACEMASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Shaw, J.P, Petsko, G.A, Ringe, D. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9-A resolution.
Biochemistry, 36, 1997
|
|
6QG9
 
 | | Crystal structure of Ideonella sakaiensis MHETase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
1S79
 
 | | Solution structure of the central RRM of human La protein | | Descriptor: | Lupus La protein | | Authors: | Alfano, C, Sanfelice, D, Babon, J, Kelly, G, Jacks, A, Curry, S, Conte, M.R. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1S9K
 
 | |
3M9Y
 
 | | Crystal structure of Triosephosphate isomerase from methicillin resistant Staphylococcus aureus at 1.9 Angstrom resolution | | Descriptor: | CITRIC ACID, SODIUM ION, Triosephosphate isomerase | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-03-23 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
1S9Q
 
 | | crystal structure of the ligand-binding domain of the estrogen-related receptor gamma in complex with 4-hydroxytamoxifen | | Descriptor: | 4-HYDROXYTAMOXIFEN, CHOLIC ACID, Estrogen-related receptor gamma | | Authors: | Greschik, H, Flaig, R, Renaud, J.P, Moras, D. | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Deactivation of the Estrogen-related Receptor {gamma} by Diethylstilbestrol or 4-Hydroxytamoxifen and Determinants of Selectivity.
J.Biol.Chem., 279, 2004
|
|
