5TPL
 
 | |
5O1M
 
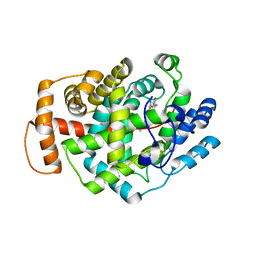 | | Structure of Latex Clearing Protein LCP in the closed state | | 分子名称: | 1,2-ETHANEDIOL, PROTOPORPHYRIN IX CONTAINING FE, Rubber oxygenase | | 著者 | Ilcu, L, Roether, W, Birke, J, Brausemann, A, Einsle, O, Jendrossek, D. | | 登録日 | 2017-05-18 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and Functional Analysis of Latex Clearing Protein (Lcp) Provides Insight into the Enzymatic Cleavage of Rubber.
Sci Rep, 7, 2017
|
|
5TX1
 
 | | Cryo-Electron microscopy structure of species-D human adenovirus 26 | | 分子名称: | Fiber, Hexon protein, PIIIa, ... | | 著者 | Reddy, V, Yu, X, Veesler, D. | | 登録日 | 2016-11-15 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of human adenovirus D26 reveals the conservation of structural organization among human adenoviruses.
Sci Adv, 3, 2017
|
|
5O3N
 
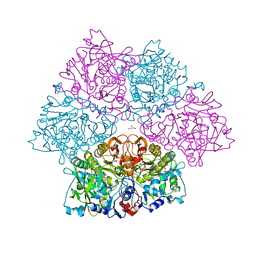 | | Crystal structure of E. cloacae 3,4-dihydroxybenzoic acid decarboxylase (AroY) reconstituted with prFMN | | 分子名称: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3,4-dihydroxybenzoate decarboxylase, GLYCEROL, ... | | 著者 | Marshall, S.A, Leys, D. | | 登録日 | 2017-05-24 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O3Z
 
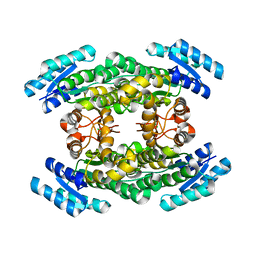 | | Crystal structure of Sorbitol-6-Phosphate 2-dehydrogenase SrlD from Erwinia amylovora | | 分子名称: | CHLORIDE ION, Sorbitol-6-phosphate dehydrogenase | | 著者 | Salomone-Stagni, M, Bartho, J.D, Bellini, D, Walsh, M.A, Benini, S. | | 登録日 | 2017-05-25 | | 公開日 | 2018-06-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural and functional analysis of Erwinia amylovora SrlD. The first crystal structure of a sorbitol-6-phosphate 2-dehydrogenase.
J.Struct.Biol., 203, 2018
|
|
5O5J
 
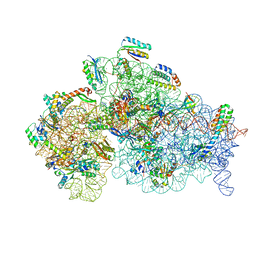 | | Structure of the 30S small ribosomal subunit from Mycobacterium smegmatis | | 分子名称: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | 著者 | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | 登録日 | 2017-06-02 | | 公開日 | 2017-07-12 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.451 Å) | | 主引用文献 | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
5O8B
 
 | | Difference-refined excited-state structure of rsEGFP2 1ps following 400nm-laser irradiation of the off-state. | | 分子名称: | Green fluorescent protein | | 著者 | Coquelle, N, Sliwa, M, Woodhouse, J, Schiro, G, Adam, V, Aquila, A, Barends, T.R.M, Boutet, S, Byrdin, M, Carbajo, S, De la Mora, E, Doak, R.B, Feliks, M, Fieschi, F, Foucar, L, Guillon, V, Hilpert, M, Hunter, M, Jakobs, S, Koglin, J.E, Kovacsova, G, Lane, T.J, Levy, B, Liang, M, Nass, K, Ridard, J, Robinson, J.S, Roome, C.M, Ruckebusch, C, Seaberg, M, Thepaut, M, Cammarata, M, Demachy, I, Field, M, Shoeman, R.L, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | 登録日 | 2017-06-12 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Chromophore twisting in the excited state of a photoswitchable fluorescent protein captured by time-resolved serial femtosecond crystallography.
Nat Chem, 10, 2018
|
|
5TUN
 
 | | Crystal structure of uninhibited human Cathepsin K at 1.62 Angstrom resolution | | 分子名称: | Cathepsin K | | 著者 | Aguda, A.H, Kruglyak, N, Nguyen, N.T, Law, S, Bromme, D, Brayer, G.D. | | 登録日 | 2016-11-06 | | 公開日 | 2017-01-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Identification of mouse cathepsin K structural elements that regulate the potency of odanacatib.
Biochem. J., 474, 2017
|
|
5NY5
 
 | | The apo structure of 3,4-dihydroxybenzoic acid decarboxylases from Enterobacter cloacae | | 分子名称: | 3,4-dihydroxybenzoate decarboxylase, GLYCEROL | | 著者 | Dordic, A, Gruber, K, Payer, S, Glueck, S, Pavkov-Keller, T, Marshall, S, Leys, D. | | 登録日 | 2017-05-11 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5UA7
 
 | | Ocellatin-LB2, solution structure in SDS micelle by NMR spectroscopy | | 分子名称: | Ocellatin-LB2 | | 著者 | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | 登録日 | 2016-12-19 | | 公開日 | 2017-03-29 | | 最終更新日 | 2018-04-18 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5OC5
 
 | |
5OCB
 
 | | Crystal structure of nitric oxide bound D97N mutant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii | | 分子名称: | COPPER (II) ION, HEME C, NITRIC OXIDE, ... | | 著者 | Dong, J, Sasaki, D, Eady, R, Antonyuk, S.V, Hasnain, S.S. | | 登録日 | 2017-06-30 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Activation of redox tyrosine switch is required for ligand binding at the catalytic site in heme-cu nitrite reductases
To be published
|
|
5U9D
 
 | | Discovery of a potent BTK inhibitor with a novel binding mode using parallel selections with a DNA-encoded chemical library | | 分子名称: | (R)-N-methyl-2-(3-((quinoxalin-6-ylamino)methyl)furan-2-carbonyl)-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxamide, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Cuozzo, J.W, Centrella, P.A, Gikunju, D, Habeshian, S, Hupp, C.D, Keefe, A.D, Sigel, E, Soutter, H.H, Thomson, H.A, Zhang, Y, Clark, M.A. | | 登録日 | 2016-12-16 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Discovery of a Potent BTK Inhibitor with a Novel Binding Mode by Using Parallel Selections with a DNA-Encoded Chemical Library.
Chembiochem, 18, 2017
|
|
5OC0
 
 | | Structure of E. coli superoxide oxidase | | 分子名称: | Cytochrome b561, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Lundgren, C.A.K, Sjostrand, D, Biner, O, Bennett, M, von Ballmoos, C, Hogbom, M. | | 登録日 | 2017-06-29 | | 公開日 | 2018-06-20 | | 最終更新日 | 2018-11-07 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Scavenging of superoxide by a membrane-bound superoxide oxidase.
Nat. Chem. Biol., 14, 2018
|
|
5U9Q
 
 | | Ocellatin-LB1 | | 分子名称: | Ocellatin-LB1 | | 著者 | Gusmao, K.A.G, Santos, D.M, de Lima, M.E, Pilo-Veloso, D, Resende, J.M. | | 登録日 | 2016-12-17 | | 公開日 | 2017-12-13 | | 最終更新日 | 2018-04-18 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5U9V
 
 | | Ocellatin-LB1, solution structure in DPC micelle by NMR spectroscopy | | 分子名称: | Ocellatin-LB1 | | 著者 | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | 登録日 | 2016-12-18 | | 公開日 | 2017-03-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Ocellatin-LB1
To Be Published
|
|
5OD0
 
 | | Crystal structure of ACPA E4 | | 分子名称: | Fab fragment of ACPA E4 - Light chain, Fab fragment of ACPA E4 - heavy chain, GLYCEROL | | 著者 | Dobritzsch, D, Ge, C, Holmdahl, R. | | 登録日 | 2017-07-04 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
5UBF
 
 | |
5OF7
 
 | | Cu nitrite reductase serial data at varying temperatures 190K 0.48MGy | | 分子名称: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | 登録日 | 2017-07-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5OFF
 
 | | Cu nitrite reductase serial data at varying temperatures RT 0.03MGy | | 分子名称: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | 登録日 | 2017-07-10 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5OG4
 
 | | Cu nitrite reductase serial data at varying temperatures RT 0.18MGy | | 分子名称: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | 登録日 | 2017-07-11 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
5NVK
 
 | | Crystal structure of the human 4EHP-GIGYF1 complex | | 分子名称: | Eukaryotic translation initiation factor 4E type 2, GRB10-interacting GYF protein 1 | | 著者 | Peter, D, Valkov, E. | | 登録日 | 2017-05-04 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
5NX5
 
 | | Crystal structure of Linalool/Nerolidol synthase from Streptomyces clavuligerus in complex with 2-fluorogeranyl diphosphate | | 分子名称: | (2Z)-2-fluoro-3,7-dimethylocta-2,6-dien-1-yl trihydrogen diphosphate, CHLORIDE ION, GLYCEROL, ... | | 著者 | Karuppiah, V, Leys, D, Scrutton, N.S. | | 登録日 | 2017-05-09 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural Basis of Catalysis in the Bacterial Monoterpene Synthases Linalool Synthase and 1,8-Cineole Synthase.
ACS Catal, 7, 2017
|
|
5S81
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010947a | | 分子名称: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | 著者 | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | 登録日 | 2020-12-11 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | XChem group deposition
To Be Published
|
|
5S7R
 
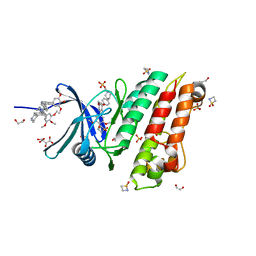 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010918a | | 分子名称: | 1,2-ETHANEDIOL, 1lambda~6~,2-thiazetidine-1,1-dione, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | 著者 | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | 登録日 | 2020-12-11 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | XChem group deposition
To Be Published
|
|
