1FF5
 
 | | STRUCTURE OF E-CADHERIN DOUBLE DOMAIN | | 分子名称: | CALCIUM ION, EPITHELIAL CADHERIN | | 著者 | Pertz, O, Bozic, D, Koch, A.W, Fauser, C, Brancaccio, A, Engel, J. | | 登録日 | 2000-07-25 | | 公開日 | 2000-08-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | A new crystal structure, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation.
EMBO J., 18, 1999
|
|
6EI5
 
 | |
1PQJ
 
 | | T4 LYSOZYME CORE REPACKING MUTANT A111V/CORE10/TA | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | 著者 | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | 登録日 | 2003-06-18 | | 公開日 | 2003-10-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | REPACKING THE CORE OF T4 LYSOZYME BY AUTOMATED DESIGN
J.Mol.Biol., 332, 2003
|
|
6CGY
 
 | | Structure of the Quorum Quenching lactonase from Alicyclobacillus acidoterrestris bound to a phosphate anion | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, COBALT (II) ION, ... | | 著者 | Bergonzi, C, Schwab, M, Naik, T, Daude, D, Chabriere, E, Elias, M. | | 登録日 | 2018-02-21 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and Biochemical Characterization of AaL, a Quorum Quenching Lactonase with Unusual Kinetic Properties.
Sci Rep, 8, 2018
|
|
3M7R
 
 | | Crystal structure of VDR H305Q mutant | | 分子名称: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Vitamin D3 receptor | | 著者 | Rochel, N, Hourai, S, Moras, D, Structural Proteomics in Europe (SPINE) | | 登録日 | 2010-03-17 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of hereditary vitamin D-resistant rickets--associated mutant H305Q of vitamin D nuclear receptor bound to its natural ligand
J.Steroid Biochem.Mol.Biol., 121, 2010
|
|
6PVK
 
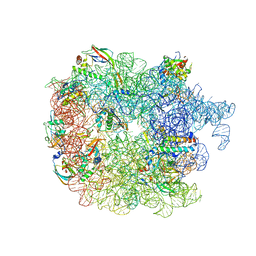 | | Bacterial 45SRbgA ribosomal particle class A | | 分子名称: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | 著者 | Ortega, J, Seffouh, A, Jain, N, Jahagirdar, D, Basu, K, Razi, A, Ni, X, Guarne, A, Britton, R.A. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
1Q40
 
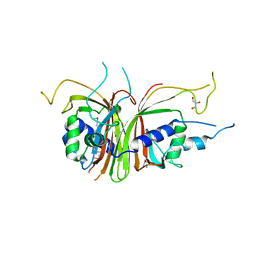 | | Crystal structure of the C. albicans Mtr2-Mex67 M domain complex | | 分子名称: | GLYCEROL, MRNA TRANSPORT REGULATOR Mtr2, mRNA export factor MEX67 | | 著者 | Senay, C, Ferrari, P, Rocher, C, Rieger, K.J, Winter, J, Platel, D, Bourne, Y. | | 登録日 | 2003-08-01 | | 公開日 | 2003-12-09 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The mtr2-mex67 ntf2-like domain complex: Structural
insights into a dual role of MTR2 for yeast nuclear export
J.Biol.Chem., 278, 2003
|
|
3M9Q
 
 | |
3M9Z
 
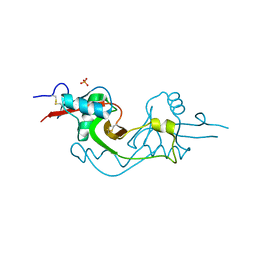 | | Crystal Structure of extracellular domain of mouse NKR-P1A | | 分子名称: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | 著者 | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | 登録日 | 2010-03-23 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Molecular architecture of mouse activating NKR-P1 receptors.
J.Struct.Biol., 175, 2011
|
|
6UCT
 
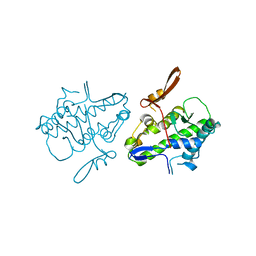 | | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant) | | 分子名称: | p9-1 | | 著者 | Llauger, G, Klinke, S, Monti, D, Sycz, G, Cerutti, M.L, Goldbaum, F.A, del Vas, M, Otero, L.H. | | 登録日 | 2019-09-17 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.47 Å) | | 主引用文献 | Crystal structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (C-arm deletion mutant)
To Be Published
|
|
1PTX
 
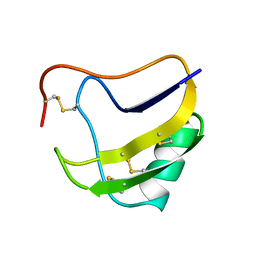 | |
1FQE
 
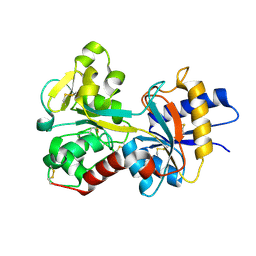 | | CRYSTAL STRUCTURES OF MUTANT (K206A) THAT ABOLISH THE DILYSINE INTERACTION IN THE N-LOBE OF HUMAN TRANSFERRIN | | 分子名称: | CARBONATE ION, FE (III) ION, POTASSIUM ION, ... | | 著者 | Nurizzo, D, Baker, H.M, Baker, E.N. | | 登録日 | 2000-09-04 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures and iron release properties of mutants (K206A and K296A) that abolish the dilysine interaction in the N-lobe of human transferrin.
Biochemistry, 40, 2001
|
|
6UD0
 
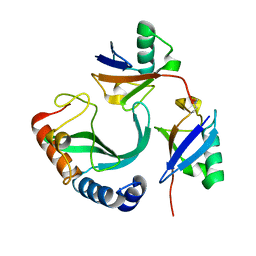 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | 分子名称: | Tumor susceptibility gene 101 protein, Ubiquitin | | 著者 | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | 登録日 | 2019-09-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
6UG1
 
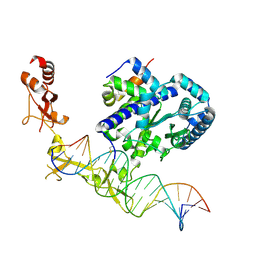 | |
3MBS
 
 | | Crystal structure of 8mer PNA | | 分子名称: | 1,2-ETHANEDIOL, Peptide Nucleic Acid | | 著者 | Yeh, J.I, Pohl, E, Truan, D, He, W, Sheldrick, G.M, Achim, C. | | 登録日 | 2010-03-26 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | The crystal structure of non-modified and bipyridine-modified PNA duplexes.
Chemistry, 16, 2010
|
|
1PVH
 
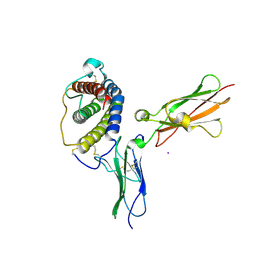 | | Crystal structure of leukemia inhibitory factor in complex with gp130 | | 分子名称: | IODIDE ION, Interleukin-6 receptor beta chain, Leukemia inhibitory factor | | 著者 | Boulanger, M.J, Bankovich, A.J, Kortemme, T, Baker, D, Garcia, K.C. | | 登録日 | 2003-06-27 | | 公開日 | 2003-10-14 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130.
Mol.Cell, 12, 2003
|
|
1POK
 
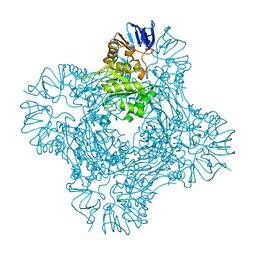 | | Crystal structure of Isoaspartyl Dipeptidase | | 分子名称: | ASPARAGINE, Isoaspartyl dipeptidase, SULFATE ION, ... | | 著者 | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | 登録日 | 2003-06-15 | | 公開日 | 2004-06-22 | | 最終更新日 | 2018-05-30 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
1FI9
 
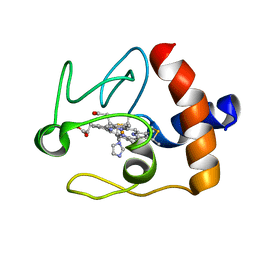 | | SOLUTION STRUCTURE OF THE IMIDAZOLE COMPLEX OF CYTOCHROME C | | 分子名称: | CYTOCHROME C, HEME C, IMIDAZOLE | | 著者 | Banci, L, Bertini, I, Liu, G, Lu, J, Reddig, T, Tang, W, Wu, Y, Zhu, D. | | 登録日 | 2000-08-03 | | 公開日 | 2000-08-23 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c
J.Biol.Inorg.Chem., 6, 2001
|
|
6VA7
 
 | |
1Q2L
 
 | |
6CJK
 
 | | Anti HIV Fab 10A | | 分子名称: | ACETATE ION, GLYCEROL, Immunoglobulin Fab heavy chain, ... | | 著者 | Hangartner, L, Ward, A.B, Wilson, I.A, Oyen, D. | | 登録日 | 2018-02-26 | | 公開日 | 2018-08-15 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.795 Å) | | 主引用文献 | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
3MGU
 
 | | Structure of S. cerevisiae Tpa1 protein, a proline hydroxylase modifying ribosomal protein Rps23 | | 分子名称: | FE (II) ION, PKHD-type hydroxylase TPA1 | | 著者 | Henri, J, Rispal, D, Bayart, E, van Tilbeurgh, H, Seraphin, B, Graille, M. | | 登録日 | 2010-04-07 | | 公開日 | 2010-07-14 | | 最終更新日 | 2018-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and functional insights into S. cerevisiae Tpa1, a putative prolyl hydroxylase influencing translation termination and transcription
To be Published
|
|
6Q6G
 
 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | 分子名称: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | 著者 | Zhang, S, Barford, D. | | 登録日 | 2018-12-11 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
1Q42
 
 | | Crystal structure analysis of the Candida albicans Mtr2 | | 分子名称: | MRNA TRANSPORT REGULATOR Mtr2 | | 著者 | Senay, C, Ferrari, P, Rocher, C, Rieger, K.J, Winter, J, Platel, D, Bourne, Y. | | 登録日 | 2003-08-01 | | 公開日 | 2003-12-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The mtr2-mex67 ntf2-like domain complex: Structural
insights into a dual role of MTR2 for yeast nuclear export
J.Biol.Chem., 278, 2003
|
|
1FPY
 
 | |
