8GA6
 
 | |
8GA7
 
 | |
2Y68
 
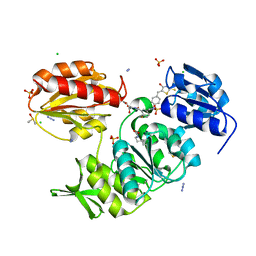 | | Structure-based design of a new series of D-glutamic acid-based inhibitors of bacterial MurD ligase | | 分子名称: | 2-[[2-fluoro-5-[[[4-[(Z)-(4-oxo-2-sulfanylidene-1,3-thiazolidin-5-ylidene)methyl]phenyl]amino]methyl]phenyl]carbonylamino]pentanedioic acid, AZIDE ION, CHLORIDE ION, ... | | 著者 | Tomasic, T, Zidar, N, Sink, R, Kovac, A, Patin, D, Blanot, D, Contreras-Martel, C, Dessen, A, Muller-Premru, M, Zega, A, Gobec, S, Peterlin-Masic, L, Kikelj, D. | | 登録日 | 2011-01-20 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Structure-based design of a new series of D-glutamic acid based inhibitors of bacterial UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase (MurD).
J. Med. Chem., 54, 2011
|
|
2BMC
 
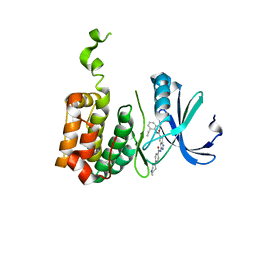 | | Aurora-2 T287D T288D complexed with PHA-680632 | | 分子名称: | (3E)-N-(2,6-DIETHYLPHENYL)-3-{[4-(4-METHYLPIPERAZIN-1-YL)BENZOYL]IMINO}PYRROLO[3,4-C]PYRAZOLE-5(3H)-CARBOXAMIDE, SERINE THREONINE-PROTEIN KINASE 6 | | 著者 | Cameron, A.D, Izzo, G, Sagliano, A, Rusconi, L, Storici, P, Fancelli, D, Berta, D, Bindi, S, Catana, C, Forte, B, Giordano, P, Mantegani, S, Meroni, M, Moll, J, Pittala, V, Severino, D, Tonani, R, Varasi, M, Vulpetti, A, Vianello, P. | | 登録日 | 2005-03-11 | | 公開日 | 2005-03-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Potent and Selective Aurora Inhibitors Identified by the Expansion of a Novel Scaffold for Protein Kinase Inhibition.
J.Med.Chem., 48, 2005
|
|
1ATI
 
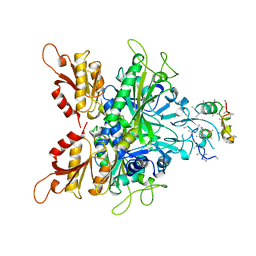 | | CRYSTAL STRUCTURE OF GLYCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | 分子名称: | GLYCYL-TRNA SYNTHETASE, GLYCYL-tRNA SYNTHETASE | | 著者 | Logan, D.T, Mazauric, M.-H, Kern, D, Moras, D. | | 登録日 | 1996-04-23 | | 公開日 | 1997-07-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal structure of glycyl-tRNA synthetase from Thermus thermophilus.
EMBO J., 14, 1995
|
|
6Y1C
 
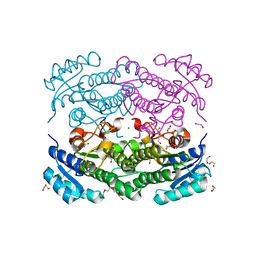 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant D54F | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | 登録日 | 2020-02-11 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6Y0Z
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q126K | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | 著者 | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | 登録日 | 2020-02-10 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6Y10
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q126H | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | 著者 | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | 登録日 | 2020-02-10 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
6WPT
 
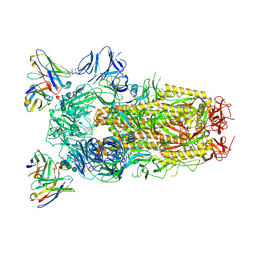 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the S309 neutralizing antibody Fab fragment (open state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 neutralizing antibody heavy chain, ... | | 著者 | Pinto, D, Park, Y.J, Beltramello, M, Walls, A.C, Tortorici, M.A, Bianchi, S, Jaconi, S, Culap, K, Zatta, F, De Marco, A, Peter, A, Guarino, B, Spreafico, R, Cameroni, E, Case, J.B, Chen, R.E, Havenar-Daughton, C, Snell, G, Virgin, H.W, Lanzavecchia, A, Diamond, M.S, Fink, K, Veesler, D, Corti, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2020-04-27 | | 公開日 | 2020-05-27 | | 最終更新日 | 2021-05-19 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody.
Nature, 583, 2020
|
|
3CCN
 
 | | X-ray structure of c-Met with triazolopyridazine inhibitor. | | 分子名称: | 4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]phenol, Hepatocyte growth factor receptor | | 著者 | Abrecht, B.K, Harmange, J.-C, Bauer, D, Dussault, I, long, A, Bellon, S.F. | | 登録日 | 2008-02-26 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
6TK1
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 20ms structure of KR2 with extrapolated, light and dark datasets | | 分子名称: | EICOSANE, RETINAL, SODIUM ION, ... | | 著者 | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | 登録日 | 2019-11-28 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TK7
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: Dark structure in acidic conditions | | 分子名称: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | 著者 | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | 登録日 | 2019-11-28 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
6TK5
 
 | | Femtosecond to millisecond structural changes in a light-driven sodium pump: 800fs+2ps structure of KR2 with extrapolated, light and dark datasets | | 分子名称: | EICOSANE, RETINAL, Sodium pumping rhodopsin | | 著者 | Skopintsev, P, Ehrenberg, D, Weinert, T, James, D, Kar, R, Johnson, P, Ozerov, D, Furrer, A, Martiel, I, Dworkowski, F, Nass, K, Knopp, G, Cirelli, C, Gashi, D, Mous, S, Wranik, M, Gruhl, T, Kekilli, D, Bruenle, S, Deupi, X, Schertler, G.F.X, Benoit, R, Panneels, V, Nogly, P, Schapiro, I, Milne, C, Heberle, J, Standfuss, J. | | 登録日 | 2019-11-28 | | 公開日 | 2020-05-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Femtosecond-to-millisecond structural changes in a light-driven sodium pump.
Nature, 583, 2020
|
|
5HLP
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH BRD3937 | | 分子名称: | 4-(2-methoxyphenyl)-3,7,7-trimethyl-1,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | 著者 | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | 登録日 | 2016-01-15 | | 公開日 | 2016-05-25 | | 最終更新日 | 2016-07-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
7PB9
 
 | | Crystal structure of tandem WH domains of Vps25 from Odinarchaeota | | 分子名称: | Tandem WH domains of Vps25 | | 著者 | Salzer, R, Bellini, D, Papatziamou, D, Robinson, N.P, Lowe, J. | | 登録日 | 2021-08-01 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Asgard archaea shed light on the evolutionary origins of the eukaryotic ubiquitin-ESCRT machinery.
Nat Commun, 13, 2022
|
|
6Y0S
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant T102E | | 分子名称: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | 著者 | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | 登録日 | 2020-02-10 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
6N44
 
 | |
5HLN
 
 | | X-RAY CRYSTAL STRUCTURE OF GSK3B IN COMPLEX WITH CHIR99021 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHIR99021, Glycogen synthase kinase-3 beta, ... | | 著者 | White, A, Lakshminarasimhan, D, Nadupalli, A, Suto, R.K. | | 登録日 | 2016-01-15 | | 公開日 | 2016-05-25 | | 最終更新日 | 2016-07-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Inhibitors of Glycogen Synthase Kinase 3 with Exquisite Kinome-Wide Selectivity and Their Functional Effects.
Acs Chem.Biol., 11, 2016
|
|
6Y15
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant T102E_Q126K | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | 著者 | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | 登録日 | 2020-02-11 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
6Y1B
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant K32A_Q126K | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | 著者 | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | 登録日 | 2020-02-11 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
2BAX
 
 | | Atomic Resolution Structure of the Double Mutant (K53,56M) of Bovine Pancreatic Phospholipase A2 | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | 著者 | Sekar, K, Yogavel, M, Velmurugan, D, Dauter, Z, Dauter, M, Tsai, M.D. | | 登録日 | 2005-10-15 | | 公開日 | 2005-10-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Atomic resolution (0.97 A) structure of the triple mutant (K53,56,121M) of bovine pancreatic phospholipase A2.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2B96
 
 | | Third Calcium ion found in an inhibitor bound phospholipase A2 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-METHOXYBENZOIC ACID, CALCIUM ION, ... | | 著者 | Sekar, K, Velmurugan, D, Yamane, T, Tsai, M.D. | | 登録日 | 2005-10-11 | | 公開日 | 2006-03-28 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Third Calcium ion found in an inhibitor bound phospholipase A2
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2EXJ
 
 | | Structure of the family43 beta-Xylosidase D128G mutant from geobacillus stearothermophilus in complex with xylobiose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | 著者 | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | 登録日 | 2005-11-08 | | 公開日 | 2006-04-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
1VRU
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | 分子名称: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | 登録日 | 1995-04-19 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1VRT
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | 分子名称: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | 著者 | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | 登録日 | 1995-04-19 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
