6BEB
 
 | | Crystal structure of VACV D13 in complex with Rifamycin SV | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, Scaffold protein D13, ... | | 著者 | Garriga, D, Accurso, C, Coulibaly, F. | | 登録日 | 2017-10-25 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural basis for the inhibition of poxvirus assembly by the antibiotic rifampicin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6IBN
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 32.8 kGy structure | | 分子名称: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | 登録日 | 2018-11-30 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
6I1F
 
 | | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with amoxicillin | | 分子名称: | 2-{1-[2-AMINO-2-(4-HYDROXY-PHENYL)-ACETYLAMINO]-2-OXO-ETHYL}-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Penicillin-binding protein,Penicillin-binding protein | | 著者 | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | 登録日 | 2018-10-28 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with amoxicillin
To Be Published
|
|
6BDO
 
 | |
6BEH
 
 | | Crystal structure of VACV D13 in complex with Rifapentine | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, RIFAPENTINE, ... | | 著者 | Garriga, D, Accurso, C, Coulibaly, F. | | 登録日 | 2017-10-25 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for the inhibition of poxvirus assembly by the antibiotic rifampicin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1KYN
 
 | | Cathepsin-G | | 分子名称: | (2-NAPHTHALEN-2-YL-1-NAPHTHALEN-1-YL-2-OXO-ETHYL)-PHOSPHONIC ACID, cathepsin G | | 著者 | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond Jr, H.R, Corcoran, T.W, De Garavilla, L, Kauffman, J.A, Recacha, R, Chattopadhyay, D, Andrade-Gordon, P, Maryanoff, B.E. | | 登録日 | 2002-02-05 | | 公開日 | 2002-05-01 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Nonpeptide inhibitors of cathepsin G: optimization of a novel beta-ketophosphonic acid lead by structure-based drug design.
J.Am.Chem.Soc., 124, 2002
|
|
6ZAS
 
 | | Damage-free as-isolated copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) determined by serial femtosecond rotation crystallography | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | 著者 | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | 登録日 | 2020-06-05 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
1L5I
 
 | | 30-CONFORMER NMR ENSEMBLE OF THE N-TERMINAL, DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN FROM A GEMINIVIRUS (TOMATO YELLOW LEAF CURL VIRUS-SARDINIA) | | 分子名称: | Rep protein | | 著者 | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | 登録日 | 2002-03-07 | | 公開日 | 2002-09-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6AVU
 
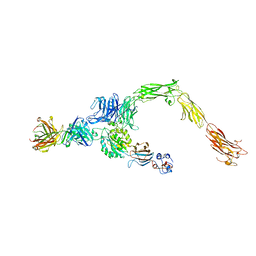 | | Human alpha-V beta-3 Integrin (open conformation) in complex with the therapeutic antibody LM609 | | 分子名称: | Fab LM609 heavy chain, Fab LM609 light chain, Integrin alpha-V, ... | | 著者 | Borst, A.J, James, Z.N, Zagotta, W.N, Ginsberg, M, Rey, F.A, DiMaio, F, Backovic, M, Veesler, D. | | 登録日 | 2017-09-04 | | 公開日 | 2017-11-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (35 Å) | | 主引用文献 | The Therapeutic Antibody LM609 Selectively Inhibits Ligand Binding to Human alpha V beta 3 Integrin via Steric Hindrance.
Structure, 25, 2017
|
|
6I1R
 
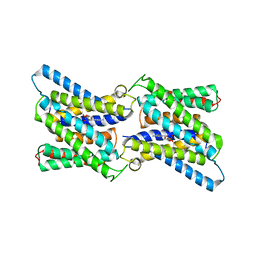 | | Crystal structure of CMP bound CST in an outward facing conformation | | 分子名称: | CMP-sialic acid transporter 1, CYTIDINE-5'-MONOPHOSPHATE | | 著者 | Nji, E, Gulati, A, Qureshi, A.A, Drew, D. | | 登録日 | 2018-10-30 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for the delivery of activated sialic acid into Golgi for sialyation.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Z97
 
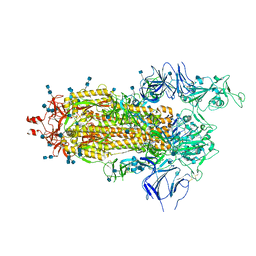 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | 著者 | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | 登録日 | 2020-06-03 | | 公開日 | 2020-07-01 | | 最終更新日 | 2020-09-23 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
1L2M
 
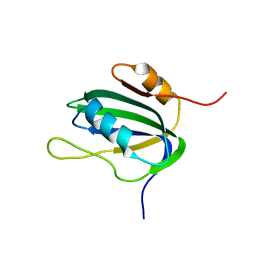 | | Minimized Average Structure of the N-terminal, DNA-binding domain of the replication initiation protein from a geminivirus (Tomato yellow leaf curl virus-Sardinia) | | 分子名称: | Rep protein | | 著者 | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | 登録日 | 2002-02-22 | | 公開日 | 2002-09-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6I54
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 2. | | 分子名称: | Influenza virus nucleoprotein, Nucleoprotein | | 著者 | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | 登録日 | 2018-11-12 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
6ZAT
 
 | | Nitrite-bound copper nitrite reductase from Bradyrhizobium sp. ORS 375 (two-domain) at 1.0 A resolution (unrestrained full matrix refinement by SHELX) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | 著者 | Rose, S.L, Antonyuk, S.V, Sasaki, D, Yamashita, K, Hirata, K, Ueno, G, Ago, H, Eady, R.R, Tosha, T, Yamamoto, M, Hasnain, S.S. | | 登録日 | 2020-06-05 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | An unprecedented insight into the catalytic mechanism of copper nitrite reductase from atomic-resolution and damage-free structures.
Sci Adv, 7, 2021
|
|
6I1G
 
 | |
6ZHJ
 
 | | 3D electron diffraction structure of thermolysin from Bacillus thermoproteolyticus | | 分子名称: | CALCIUM ION, Thermolysin, ZINC ION | | 著者 | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | 登録日 | 2020-06-23 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (3.26 Å) | | 主引用文献 | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ZHN
 
 | | 3D electron diffraction structure of thaumatin from Thaumatococcus daniellii | | 分子名称: | CHLORIDE ION, Thaumatin-1 | | 著者 | Blum, T, Housset, D, Clabbers, M.T.B, van Genderen, E, Schoehn, G, Ling, W.L, Abrahams, J.P. | | 登録日 | 2020-06-23 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.76 Å) | | 主引用文献 | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6BBN
 
 | |
6ZDH
 
 | | SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-14 | | 公開日 | 2020-07-01 | | 最終更新日 | 2021-12-22 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6I8P
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 78.4 kGy structure | | 分子名称: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | 登録日 | 2018-11-20 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
6I9C
 
 | | Structure of the OTU domain of OTULIN G281R mutant | | 分子名称: | CHLORIDE ION, GLYCEROL, Ubiquitin thioesterase otulin | | 著者 | Damgaard, R.B, Elliott, P.R, Swatek, K.N, Maher, E.R, Stepensky, P, Elpeleg, O, Komander, D, Berkun, Y. | | 登録日 | 2018-11-22 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | OTULIN deficiency in ORAS causes cell type-specific LUBAC degradation, dysregulated TNF signalling and cell death.
Embo Mol Med, 11, 2019
|
|
6ZKH
 
 | | Complex I with NADH, open1 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | 著者 | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | 登録日 | 2020-06-12 | | 公開日 | 2020-06-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6I36
 
 | | SIXTY MINUTES IRON LOADED FROG M FERRITIN | | 分子名称: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | 著者 | Mangani, S, Di Pisa, F, Pozzi, C, Turano, P, Lalli, D. | | 登録日 | 2018-11-05 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Time-Lapse Anomalous X-Ray Diffraction Shows How Fe(2+) Substrate Ions Move Through Ferritin Protein Nanocages To Oxidoreductase Sites.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6ZL3
 
 | | CRYSTAL STRUCTURE OF HRAS IN COMPLEX WITH COMPOUND 18 and GDP | | 分子名称: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Kessler, D, Fischer, G, Boettcher, J. | | 登録日 | 2020-06-30 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.031 Å) | | 主引用文献 | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
