6IKO
 
 | | Crystal structure of mouse GAS7cb | | 分子名称: | Growth arrest-specific protein 7 | | 著者 | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | 登録日 | 2018-10-16 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.756 Å) | | 主引用文献 | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
6I7H
 
 | | Crystal structure of dimeric FICD mutant K256S | | 分子名称: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION | | 著者 | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | 登録日 | 2018-11-16 | | 公開日 | 2019-09-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
7PEG
 
 | | Structure of the sporulation/germination protein YhcN from Bacillus subtilis | | 分子名称: | Probable spore germination lipoprotein YhcN | | 著者 | Liu, B, Chan, H, Bauda, E, Contreras-Martel, C, Bellard, L, Villard, A.M, Mas, C, Neumann, E, Fenel, D, Favier, A, Serrano, M, Henriques, A.O.H, Rodrigues, C.D.A, Morlot, C. | | 登録日 | 2021-08-10 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural insights into ring-building motif domains involved in bacterial sporulation.
J.Struct.Biol., 214, 2022
|
|
6I8K
 
 | | Dye type peroxidase Aa from Streptomyces lividans: 164 kGy structure | | 分子名称: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | 登録日 | 2018-11-20 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
7PKL
 
 | |
6IB2
 
 | | The structure of MKK7 in complex with the covalent 4-amino-pyrazolopyrimidine 4a | | 分子名称: | 1-[(3~{R})-3-[4-azanyl-3-[1-(4-ethanoylphenyl)-1,2,3-triazol-4-yl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Dual specificity mitogen-activated protein kinase kinase 7 | | 著者 | Wolle, P, Hardick, J, Mueller, M.P, Rauh, D. | | 登録日 | 2018-11-28 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Targeting the MKK7-JNK (Mitogen-Activated Protein Kinase Kinase 7-c-Jun N-Terminal Kinase) Pathway with Covalent Inhibitors.
J.Med.Chem., 62, 2019
|
|
1IJY
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF MOUSE FRIZZLED 8 (MFZ8) | | 分子名称: | FRIZZLED HOMOLOG 8 | | 著者 | Dann III, C.E, Hsieh, J.C, Rattner, A, Sharma, D, Nathans, J, Leahy, D.J. | | 登録日 | 2001-05-01 | | 公開日 | 2001-07-11 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains.
Nature, 412, 2001
|
|
6IAR
 
 | | Tricyclic indazoles a novel class of selective estrogen receptor degrader antagonists | | 分子名称: | 3-[4-[(6~{R})-7-(2-methylpropyl)-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]phenyl]propanoic acid, Estrogen receptor | | 著者 | Scott, J.S, Bailey, A, Buttar, D, Carbajo, R.J, Curwen, J, Davies, R.D.M, Degorce, S.L, Donald, C, Gangl, E, Greenwood, R, Groombridge, S.D, Johnson, T, Lamont, S, Lawson, M, Lister, A, Morrow, C, Moss, T, Pink, J.H, Polanski, R. | | 登録日 | 2018-11-27 | | 公開日 | 2019-01-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Tricyclic Indazoles-A Novel Class of Selective Estrogen Receptor Degrader Antagonists.
J.Med.Chem., 62, 2019
|
|
7C7S
 
 | | Cryo-EM structure of the CGP54626-bound human GABA(B) receptor in inactive state. | | 分子名称: | (R)-(cyclohexylmethyl)[(2S)-3-{[(1S)-1-(3,4-dichlorophenyl)ethyl]amino}-2-hydroxypropyl]phosphinic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid type B receptor subunit 1, ... | | 著者 | Mao, C, Shen, C, Li, C, Shen, D, Xu, C, Zhang, S, Zhou, R, Shen, Q, Chen, L, Jiang, Z, Liu, J, Zhang, Y. | | 登録日 | 2020-05-26 | | 公開日 | 2020-07-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures of inactive and active GABABreceptor.
Cell Res., 30, 2020
|
|
6IJI
 
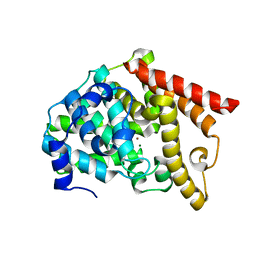 | | Crystal structure of PDE10 in complex with inhibitor 2b | | 分子名称: | 2-{2-[5-methyl-1-(pyridin-4-yl)-1H-benzimidazol-2-yl]ethyl}-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | 著者 | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | 登録日 | 2018-10-10 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | 分子名称: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | 著者 | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | 登録日 | 2020-06-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2021-04-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
6EIG
 
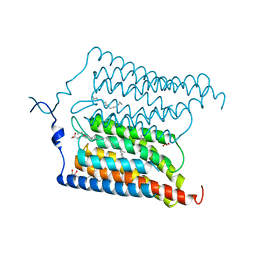 | | Crystal structure of N24Q/C128T mutant of Channelrhodopsin 2 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Archaeal-type opsin 2, EICOSANE, ... | | 著者 | Kovalev, K, Borshchevskiy, V, Volkov, O, Polovinkin, V, Marin, E, Balandin, T, Astashkin, R, Bamann, C, Bueldt, G, Willlbold, D, Popov, A, Bamberg, E, Gordeliy, V. | | 登録日 | 2017-09-19 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into ion conduction by channelrhodopsin 2.
Science, 358, 2017
|
|
6RYB
 
 | |
5IMA
 
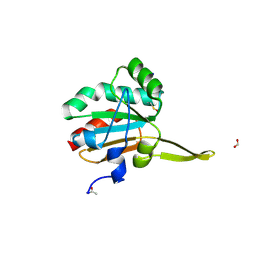 | | Xanthomonas campestris Peroxiredoxin Q - Structure F2 | | 分子名称: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | 著者 | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | 登録日 | 2016-03-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
5IMF
 
 | | Xanthomonas campestris Peroxiredoxin Q - Structure F5 | | 分子名称: | Bacterioferritin comigratory protein, FORMIC ACID, SODIUM ION | | 著者 | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | 登録日 | 2016-03-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
1IY0
 
 | | Crystal structure of the FtsH ATPase domain with AMP-PNP from Thermus thermophilus | | 分子名称: | ATP-dependent metalloprotease FtsH, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | 登録日 | 2002-07-10 | | 公開日 | 2002-11-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
5IKW
 
 | | Crystal Structure of BMP-2-inducible kinase in complex with an Indazole inhibitor | | 分子名称: | BMP-2-inducible protein kinase, N-(6-{3-[(cyclopropylsulfonyl)amino]phenyl}-1H-indazol-3-yl)cyclopropanecarboxamide | | 著者 | Counago, R.M, Sorrell, F.J, Krojer, T, Savitsky, P, Elkins, J.M, Axtman, A, Drewry, D, Wells, C, Zhang, C, Zuercher, W, Willson, T.M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Arruda, P, Gileadi, O, Structural Genomics Consortium (SGC) | | 登録日 | 2016-03-04 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal Structure of BMP-2-inducible kinase in complex with a 3-acylaminoindazole inhibitor GSK3236425A
To Be Published
|
|
7PL9
 
 | |
5IPH
 
 | | Xanthomonas campestris Peroxiredoxin Q - C84S mutant | | 分子名称: | Bacterioferritin comigratory protein, SODIUM ION | | 著者 | Perkins, A, Parsonage, D, Nelson, K.J, Poole, L.B, Karplus, A. | | 登録日 | 2016-03-09 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Peroxiredoxin Catalysis at Atomic Resolution.
Structure, 24, 2016
|
|
6I4R
 
 | | Crystal structure of the disease-causing R460G mutant of the human dihydrolipoamide dehydrogenase at 1.44 Angstrom resolution | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydrolipoyl dehydrogenase, mitochondrial, ... | | 著者 | Szabo, E, Wilk, P, Bui, D, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | 登録日 | 2018-11-10 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.439 Å) | | 主引用文献 | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
6RJN
 
 | | Crystal structure of a Fungal Catalase at 2.3 Angstroms | | 分子名称: | CHLORIDE ION, Catalase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Gomez, S, Navas-Yuste, S, Payne, A.M, Rivera, W, Lopez-Estepa, M, Brangbour, C, Fulla, D, Juanhuix, J, Fernandez, F.J, Vega, M.C. | | 登録日 | 2019-04-28 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.295 Å) | | 主引用文献 | Peroxisomal catalases from the yeasts Pichia pastoris and Kluyveromyces lactis as models for oxidative damage in higher eukaryotes.
Free Radic. Biol. Med., 141, 2019
|
|
5IQ6
 
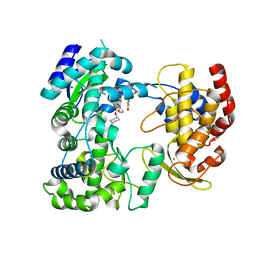 | | Crystal structure of Dengue virus serotype 3 RNA dependent RNA polymerase bound to HeE1-2Tyr, a new pyridobenzothizole inhibitor | | 分子名称: | N-[8-(cyclohexyloxy)-1-oxo-2-phenyl-1H-pyrido[2,1-b][1,3]benzothiazole-4-carbonyl]-L-tyrosine, RNA dependent RNA polymerase, ZINC ION | | 著者 | Tarantino, D, Mastrangelo, E, Milani, M. | | 登録日 | 2016-03-10 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Targeting flavivirus RNA dependent RNA polymerase through a pyridobenzothiazole inhibitor.
Antiviral Res., 134, 2016
|
|
1IJX
 
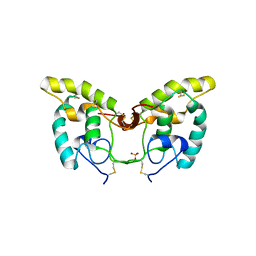 | | CRYSTAL STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF SECRETED FRIZZLED-RELATED PROTEIN 3 (SFRP-3;FZB) | | 分子名称: | SECRETED FRIZZLED-RELATED SEQUENCE PROTEIN 3, SULFATE ION | | 著者 | Dann III, C.E, Hsieh, J.C, Rattner, A, Sharma, D, Nathans, J, Leahy, D.J. | | 登録日 | 2001-04-30 | | 公開日 | 2001-07-11 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains.
Nature, 412, 2001
|
|
6I6G
 
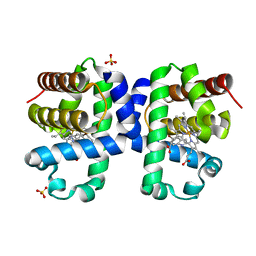 | | Dehaloperoxidase B from Amphitrite ornata - complex with 5-bromoindole | | 分子名称: | 5-bromanyl-1~{H}-indole, Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Moreno-Chicano, T, Ebrahim, A.E, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Sugimoto, H, Tono, K, Owada, S, Duyvesteyn, H. | | 登録日 | 2018-11-15 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
6I7B
 
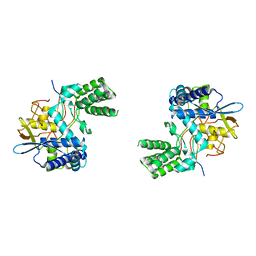 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3. | | 分子名称: | Nucleoprotein | | 著者 | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | 登録日 | 2018-11-16 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
