3ZQG
 
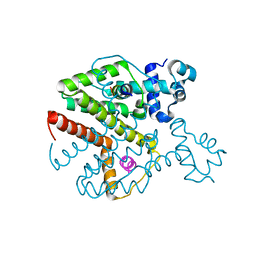 | | Structure of Tetracycline repressor in complex with antiinducer peptide-TAP2 | | 分子名称: | ANTI-INDUCER PEPTIDE TAP2, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | 著者 | Sevvana, M, Goeke, D, Stoeckle, C, Kaspar, D, Grubmueller, S, Goetz, C, Wimmer, C, Berens, C, Klotzsche, M, Muller, Y.A, Hillen, W. | | 登録日 | 2011-06-09 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | An Exclusive Alpha/Beta Code Directs Allostery in Tetr-Peptide Complexes.
J.Mol.Biol., 416, 2012
|
|
5GPE
 
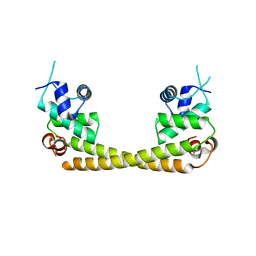 | | Crystal structure of the transcription regulator PbrR691 from Ralstonia metallidurans CH34 in complex with Lead(II) | | 分子名称: | LEAD (II) ION, Transcriptional regulator, MerR-family | | 著者 | Huang, S.Q, Chen, W.Z, Wang, D, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | 登録日 | 2016-08-01 | | 公開日 | 2016-12-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Basis for the Selective Pb(II) Recognition of Metalloregulatory Protein PbrR691
Inorg Chem, 55, 2016
|
|
3OWU
 
 | |
3TC6
 
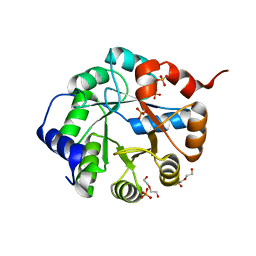 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | 分子名称: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | 著者 | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-08-08 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
3TE5
 
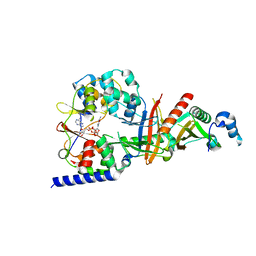 | | structure of the regulatory fragment of sacchromyces cerevisiae ampk in complex with NADH | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | 著者 | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | 登録日 | 2011-08-12 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3OZI
 
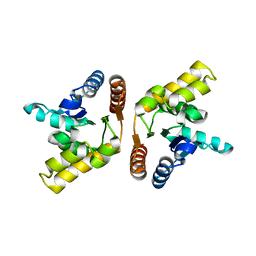 | | Crystal structure of the TIR domain from the flax disease resistance protein L6 | | 分子名称: | COBALT (II) ION, L6tr | | 著者 | Ve, T, Bernoux, M, Williams, S, Valkov, E, Warren, C, Hatters, D, Ellis, J.G, Dodds, P.N, Kobe, B. | | 登録日 | 2010-09-25 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Functional Analysis of a Plant Resistance Protein TIR Domain Reveals Interfaces for Self-Association, Signaling, and Autoregulation.
Cell Host Microbe, 9, 2011
|
|
5GSZ
 
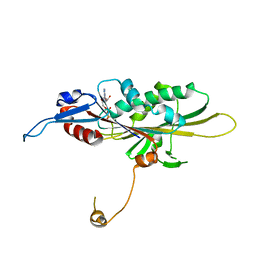 | | Crystal Structure of the KIF19A Motor Domain Complexed with Mg-ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF19, MAGNESIUM ION | | 著者 | Wang, D, Nitta, R, Hirokawa, N. | | 登録日 | 2016-08-18 | | 公開日 | 2016-09-28 | | 最終更新日 | 2020-02-26 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
3OX9
 
 | |
5H1O
 
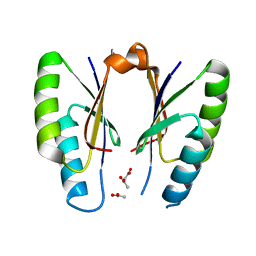 | | CRISPR-associated protein | | 分子名称: | ACETATE ION, CRISPR-associated endoribonuclease Cas2 | | 著者 | Ka, D, Jeong, U, Bae, E. | | 登録日 | 2016-10-11 | | 公開日 | 2017-10-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity
Struct Dyn, 4, 2017
|
|
5GWD
 
 | | Structure of Myroilysin | | 分子名称: | Myroilysin, ZINC ION | | 著者 | Xu, D, Ran, T, Wang, W. | | 登録日 | 2016-09-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Myroilysin Is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Is Activated by a Cysteine Switch Mechanism.
J. Biol. Chem., 292, 2017
|
|
5H2T
 
 | |
5EA3
 
 | | Crystal Structure of Inhibitor JNJ-2408068 in Complex with Prefusion RSV F Glycoprotein | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-[[2-[[1-(2-azanylethyl)piperidin-4-yl]amino]-4-methyl-benzimidazol-1-yl]methyl]-6-methyl-pyridin-3-ol, Fusion glycoprotein F0, ... | | 著者 | Battles, M.B, McLellan, J.S, Arnoult, E, Langedijk, J.P, Roymans, D. | | 登録日 | 2015-10-15 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
7K4W
 
 | | Crystal structure of Kemp Eliminase HG3.17 in the inactive state | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
5EAM
 
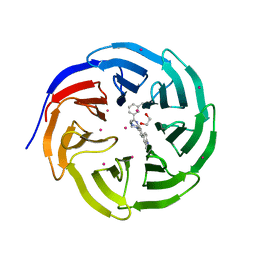 | | Crystal structure of human WDR5 in complex with compound 9o | | 分子名称: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | 著者 | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | 登録日 | 2015-10-16 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
1D0R
 
 | |
7P84
 
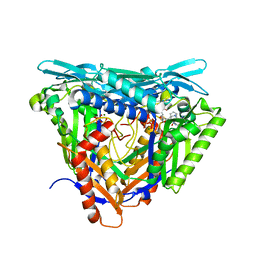 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with ONB-SAM (2-nitro benzyme S-adenosyl-methionine) | | 分子名称: | MAGNESIUM ION, S-adenosylmethionine synthase, TRIPHOSPHATE, ... | | 著者 | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | 登録日 | 2021-07-21 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.054 Å) | | 主引用文献 | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
5ELF
 
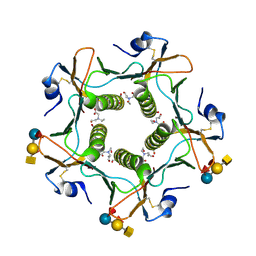 | | Cholera toxin El Tor B-pentamer in complex with A-pentasaccharide | | 分子名称: | BICINE, CALCIUM ION, Cholera enterotoxin subunit B, ... | | 著者 | Heggelund, J.E, Burschowsky, D, Krengel, U. | | 登録日 | 2015-11-04 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | High-Resolution Crystal Structures Elucidate the Molecular Basis of Cholera Blood Group Dependence.
Plos Pathog., 12, 2016
|
|
7P8M
 
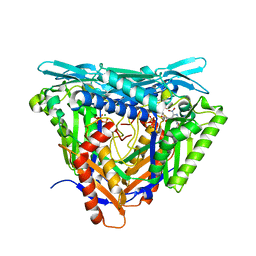 | | Crystal structure of L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii in complex with DMNB-SAM (4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine) | | 分子名称: | 4,5-dimethoxy-2-nitro benzyme S-adenosyl-methionine, MAGNESIUM ION, S-adenosylmethionine synthase, ... | | 著者 | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | 登録日 | 2021-07-23 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
5EF5
 
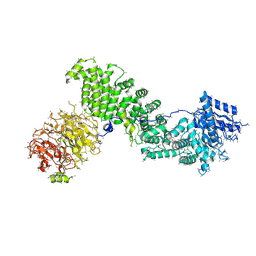 | | Crystal structure of Chaetomium thermophilum Raptor | | 分子名称: | Raptor from Chaetomium thermophilum | | 著者 | Imseng, S, Sauer, E, Aylett, C.H.S, Boehringer, D, Hall, M.N, Ban, N, Maier, T. | | 登録日 | 2015-10-23 | | 公開日 | 2015-12-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Architecture of human mTOR complex 1.
Science, 351, 2016
|
|
7P82
 
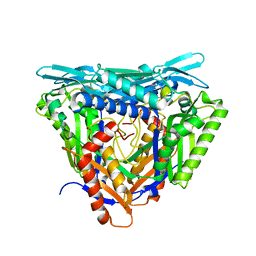 | | Crystal structure of apo form L147A/I351A variant of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii | | 分子名称: | S-adenosylmethionine synthase | | 著者 | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | 登録日 | 2021-07-21 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.042 Å) | | 主引用文献 | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
7P83
 
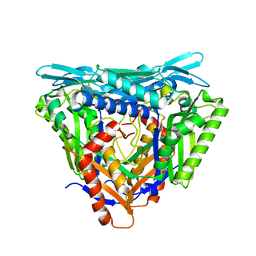 | | Crystal structure of Apo form of S-adenosylmethionine synthetase from Methanocaldococcus jannaschii | | 分子名称: | S-adenosylmethionine synthase | | 著者 | Herrmann, E, Peters, A, Cornelissen, N.V, Rentmeister, A, Kuemmel, D. | | 登録日 | 2021-07-21 | | 公開日 | 2021-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.218 Å) | | 主引用文献 | Visible-Light Removable Photocaging Groups Accepted by MjMAT Variant: Structural Basis and Compatibility with DNA and RNA Methyltransferases.
Chembiochem, 23, 2022
|
|
3TC7
 
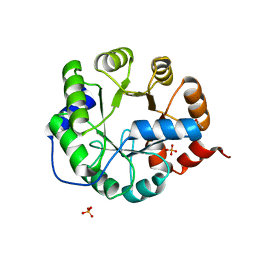 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | 分子名称: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | 著者 | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-08-08 | | 公開日 | 2011-08-24 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
1A0K
 
 | |
5EGW
 
 | |
5EPW
 
 | | C-Terminal Domain Of Human Coronavirus Nl63 Nucleocapsid Protein | | 分子名称: | Nucleoprotein | | 著者 | Szelazek, B, Kabala, W, Kus, K, Zdzalik, M, Golik, P, Florek, D, Burmistrz, M, Pyrc, K, Dubin, G. | | 登録日 | 2015-11-12 | | 公開日 | 2017-02-22 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural Characterization of Human Coronavirus NL63 N Protein.
J. Virol., 91, 2017
|
|
