4WJA
 
 | | Crystal Structure of PAXX | | 分子名称: | Uncharacterized protein C9orf142 | | 著者 | Xing, M, Yang, M, Huo, W, Feng, F, Wei, L, Ning, S, Yan, Z, Li, W, Wang, Q, Hou, M, Dong, C, Guo, R, Gao, G, Ji, J, Lan, L, Liang, H, Xu, D. | | 登録日 | 2014-09-29 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway.
Nat Commun, 6, 2015
|
|
8HUA
 
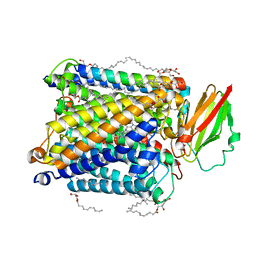 | | Serial synchrotron crystallography structure of ba3-type cytochrome c oxidase from Thermus thermophilus using a goniometer compatible flow-cell | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | 著者 | Ghosh, S, Zoric, D, Bjelcic, M, Johannesson, J, Sandelin, E, Branden, G, Neutze, R. | | 登録日 | 2022-12-22 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | A simple goniometer-compatible flow cell for serial synchrotron X-ray crystallography.
J.Appl.Crystallogr., 56, 2023
|
|
4WIQ
 
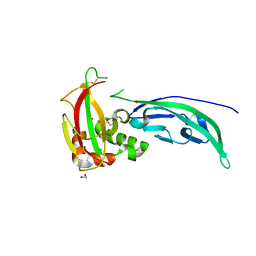 | | The structure of Murine alpha-Dystroglycan T190M mutant N-terminal domain. | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Dystroglycan | | 著者 | Brancaccio, A, Lamba, D, Cassetta, A, Bozzi, M, Covaceuszach, S, Sciandra, F, Bigotti, M.G. | | 登録日 | 2014-09-26 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | The Structure of the T190M Mutant of Murine alpha-Dystroglycan at High Resolution: Insight into the Molecular Basis of a Primary Dystroglycanopathy.
Plos One, 10, 2015
|
|
1EXX
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE INACTIVE S-ENANTIOMER BMS270395. | | 分子名称: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-2 | | 著者 | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | 登録日 | 2000-05-05 | | 公開日 | 2000-06-09 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2ZHC
 
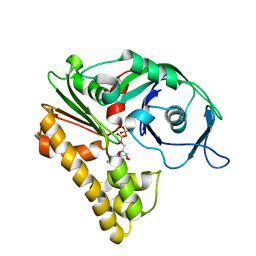 | | ParM filament | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid segregation protein parM | | 著者 | Popp, D, Narita, A, Oda, T, Fujisawa, T, Matsuo, H, Nitanai, Y, Iwasa, M, Maeda, K, Onishi, H, Maeda, Y. | | 登録日 | 2008-02-04 | | 公開日 | 2008-02-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (23 Å) | | 主引用文献 | Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability
Embo J., 27, 2008
|
|
6O6B
 
 | |
6O72
 
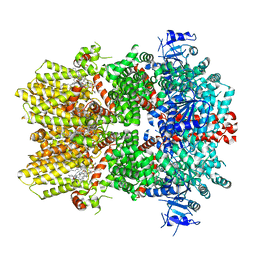 | | Structure of the TRPM8 cold receptor by single particle electron cryo-microscopy, TC-I 2014-bound state | | 分子名称: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(heptanoyloxy)methyl]ethyl octadecanoate, 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Diver, M.M, Cheng, Y, Julius, D. | | 登録日 | 2019-03-07 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural insights into TRPM8 inhibition and desensitization.
Science, 365, 2019
|
|
6O7T
 
 | | Saccharomyces cerevisiae V-ATPase Vph1-VO | | 分子名称: | Putative protein YPR170W-B, V-type proton ATPase subunit a, vacuolar isoform, ... | | 著者 | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | 登録日 | 2019-03-08 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
2ZNI
 
 | | Crystal structure of Pyrrolysyl-tRNA synthetase-tRNA(Pyl) complex from Desulfitobacterium hafniense | | 分子名称: | CALCIUM ION, Pyrrolysyl-tRNA synthetase, bacterial tRNA | | 著者 | Nozawa, K, Araiso, Y, Soll, D, Ishitani, R, Nureki, O. | | 登録日 | 2008-04-25 | | 公開日 | 2008-12-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality
Nature, 457, 2009
|
|
6O9A
 
 | | Crystal structure of MqnA complexed with 3-hydroxybenzoic acid | | 分子名称: | 3-HYDROXYBENZOIC ACID, ACETATE ION, Chorismate dehydratase | | 著者 | Hicks, K.A, Mahanta, N, Naseem, S, Fedoseyenko, D, Begley, T.P, Ealick, S.E. | | 登録日 | 2019-03-13 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.326 Å) | | 主引用文献 | Menaquinone Biosynthesis: Biochemical and Structural Studies of Chorismate Dehydratase.
Biochemistry, 58, 2019
|
|
6ODC
 
 | | Crystal structure of HDAC8 in complex with compound 30 | | 分子名称: | (2E)-3-[2-(3-cyclopentyl-5,5-dimethyl-2-oxoimidazolidin-1-yl)phenyl]-N-hydroxyprop-2-enamide, 1,2-ETHANEDIOL, Histone deacetylase 8, ... | | 著者 | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | 登録日 | 2019-03-26 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
2ZT9
 
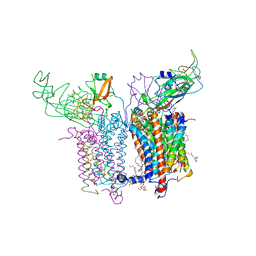 | | Crystal Structure of the Cytochrome b6f Complex from Nostoc sp. PCC 7120 | | 分子名称: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, Apocytochrome f, ... | | 著者 | Craner, W.A, Baniulis, D, Yamashita, E. | | 登録日 | 2008-09-27 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-Function, Stability, and Chemical Modification of the Cyanobacterial Cytochrome b6f Complex from Nostoc sp. PCC 7120
J.Biol.Chem., 284, 2009
|
|
6ODA
 
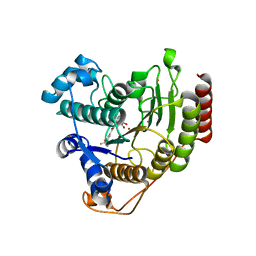 | | Crystal structure of HDAC8 in complex with compound 2 | | 分子名称: | Histone deacetylase 8, N-{2-[3-(hydroxyamino)-3-oxopropyl]phenyl}-3-(trifluoromethyl)benzamide, POTASSIUM ION, ... | | 著者 | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | 登録日 | 2019-03-26 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6OF5
 
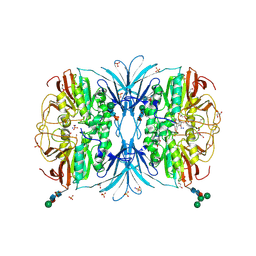 | | The crystal structure of dodecyloxy(naphthalen-1-yl)methylphosphonic acid in complex with red kidney bean purple acid phosphatase | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, ... | | 著者 | Feder, D, Schenk, G, Guddat, L.W, Hussein, W.M, McGeary, R.P. | | 登録日 | 2019-03-28 | | 公開日 | 2019-09-04 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Synthesis, evaluation and structural investigations of potent purple acid phosphatase inhibitors as drug leads for osteoporosis.
Eur.J.Med.Chem., 182, 2019
|
|
4X3J
 
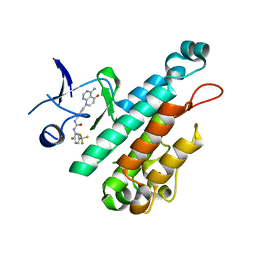 | | Selection of fragments for kinase inhibitor design: decoration is key | | 分子名称: | 1-[4-(4-amino-5-oxopyrido[2,3-d]pyrimidin-8(5H)-yl)phenyl]-3-[2-fluoro-5-(trifluoromethyl)phenyl]urea, Angiopoietin-1 receptor | | 著者 | Czodrowski, P, Hoelzemann, G, Barnickel, G, Greiner, H, Musil, D. | | 登録日 | 2014-11-30 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Selection of fragments for kinase inhibitor design: decoration is key.
J.Med.Chem., 58, 2015
|
|
6OI3
 
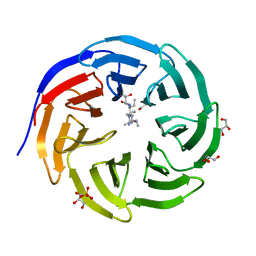 | | Crystal structure of human WDR5 in complex with monomethyl H3R2 peptide | | 分子名称: | GLYCEROL, Monomethyl H3R2 peptide, SULFATE ION, ... | | 著者 | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | 登録日 | 2019-04-08 | | 公開日 | 2020-04-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
4X4Z
 
 | |
6O7V
 
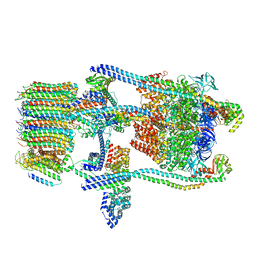 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 1 | | 分子名称: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | 登録日 | 2019-03-08 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4WKT
 
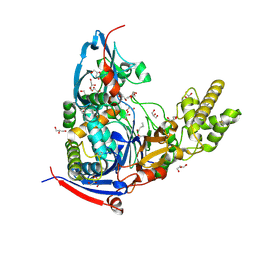 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | 分子名称: | 1-BUTANE BORONIC ACID, Acyl-homoserine lactone acylase PvdQ, GLYCEROL | | 著者 | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | 登録日 | 2014-10-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.782 Å) | | 主引用文献 | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
368D
 
 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | 分子名称: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | 著者 | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L, Malinina, L. | | 登録日 | 1997-12-19 | | 公開日 | 1998-07-15 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
3FEI
 
 | | Design and biological evaluation of novel, balanced dual PPARa/g agonists | | 分子名称: | (2S)-3-(4-{[2-(4-chlorophenyl)-1,3-thiazol-4-yl]methoxy}-2-methylphenyl)-2-ethoxypropanoic acid, Peptide motif 5 of Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | 著者 | Benz, J, Grether, U, Gsell, B, Binggeli, A, Hilpert, H, Maerki, H.P, Mohr, P, Ruf, A, Stihle, M, Schlatter, D. | | 登録日 | 2008-11-30 | | 公開日 | 2009-10-20 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Design and biological evaluation of novel, balanced dual PPARalpha/gamma agonists
Chemmedchem, 4, 2009
|
|
3A9E
 
 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | 分子名称: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | 著者 | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | 登録日 | 2009-10-24 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
3AAG
 
 | |
6OEZ
 
 | | Crystal structure of Trypanothione Reductase from Trypanosoma brucei in complex with inhibitor (+)-N-(Cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrro-lidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, N-(cyclobutylmethyl)-3-{5-[1-(pyrrolidin-1-yl)cyclohexyl]-2-(1-{[(2S)-pyrrolidin-2-yl]methyl}-1H-indol-5-yl)-1,3-thiazol-4-yl}prop-2-yn-1-amine, ... | | 著者 | Halgas, O, De Gasparo, R, Harangozo, D, Krauth-Siegel, R.L, Diederich, F, Pai, E.F. | | 登録日 | 2019-03-28 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Targeting a Large Active Site: Structure-Based Design of Nanomolar Inhibitors of Trypanosoma brucei Trypanothione Reductase.
Chemistry, 25, 2019
|
|
384D
 
 | |
