1YDR
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H7 PROTEIN KINASE INHIBITOR 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE | | 分子名称: | 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE, C-AMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR PEPTIDE | | 著者 | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | 登録日 | 1996-07-24 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDS
 
 | | Structure of CAMP-dependent protein kinase, alpha-catalytic subunit in complex with H8 protein kinase inhibitor [N-(2-methylamino)ethyl]-5-isoquinolinesulfonamide | | 分子名称: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(METHYLAMINO)ETHYL]-5-ISOQUINOLINESULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | 著者 | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | 登録日 | 1996-07-24 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDT
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H89 PROTEIN KINASE INHIBITOR N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE | | 分子名称: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | 著者 | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | 登録日 | 1996-07-24 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1Z60
 
 | | Solution structure of the carboxy-terminal domain of human TFIIH P44 subunit | | 分子名称: | TFIIH basal transcription factor complex p44 subunit, ZINC ION | | 著者 | Kellenberger, E, Dominguez, C, Fribourg, S, Wasielewski, E, Moras, D, Poterszman, A, Boelens, R, Kieffer, B. | | 登録日 | 2005-03-21 | | 公開日 | 2005-04-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C-terminal domain of TFIIH P44 subunit reveals a novel type of C4C4 ring domain involved in protein-protein interactions.
J.Biol.Chem., 280, 2005
|
|
6FKA
 
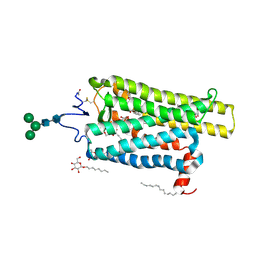 | | Crystal structure of N2C/D282C stabilized opsin bound to RS11 | | 分子名称: | (2~{S})-2-(3,4-dichlorophenyl)-3-methyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | 著者 | Mattle, D, Standfuss, J, Dawson, R. | | 登録日 | 2018-01-23 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1XS0
 
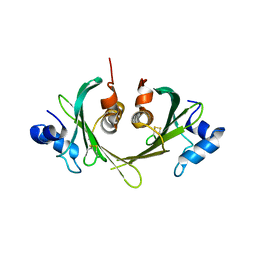 | | Structure of the E. coli Ivy protein | | 分子名称: | Inhibitor of vertebrate lysozyme | | 著者 | Abergel, C, Monchois, V, Byrn, D, Lazzaroni, J.C, Claverie, J.M. | | 登録日 | 2004-10-18 | | 公開日 | 2004-11-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Structure and evolution of the Ivy protein family, unexpected lysozyme inhibitors in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6YS5
 
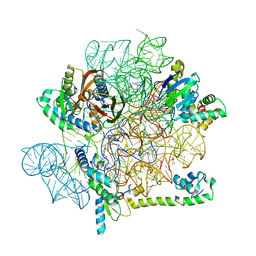 | | Acinetobacter baumannii ribosome-amikacin complex - 30S subunit head | | 分子名称: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | 著者 | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | 登録日 | 2020-04-21 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
1XO1
 
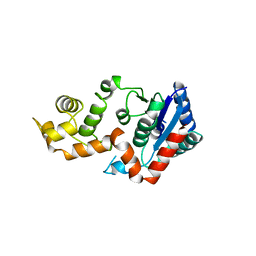 | | T5 5'-EXONUCLEASE MUTANT K83A | | 分子名称: | 5'-EXONUCLEASE | | 著者 | Ceska, T.A, Suck, D, Sayers, J.R. | | 登録日 | 1998-11-19 | | 公開日 | 1999-04-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutagenesis of conserved lysine residues in bacteriophage T5 5'-3' exonuclease suggests separate mechanisms of endo-and exonucleolytic cleavage.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
6FKC
 
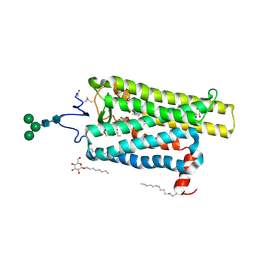 | | Crystal structure of N2C/D282C stabilized opsin bound to RS15 | | 分子名称: | 3-[1'-[(2~{S})-2-(4-chlorophenyl)-3-methyl-butanoyl]spiro[1,3-benzodioxole-2,4'-piperidine]-5-yl]propanoic acid, PALMITIC ACID, Rhodopsin, ... | | 著者 | Mattle, D, Standfuss, J, Dawson, R. | | 登録日 | 2018-01-23 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FEL
 
 | |
6FK8
 
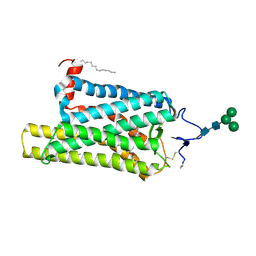 | | Crystal structure of N2C/D282C stabilized opsin bound to RS08 | | 分子名称: | (2~{R},3~{S})-3-azanyl-2-(4-chlorophenyl)-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | 著者 | Mattle, D, Standfuss, J, Dawson, R. | | 登録日 | 2018-01-23 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FK7
 
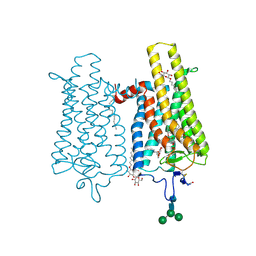 | | Crystal structure of N2C/D282C stabilized opsin bound to RS06 | | 分子名称: | (2~{R},3~{R})-2-(4-chlorophenyl)-3-oxidanyl-1-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-butan-1-one, PALMITIC ACID, Rhodopsin, ... | | 著者 | Mattle, D, Standfuss, J, Dawson, R. | | 登録日 | 2018-01-23 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FKD
 
 | | Crystal structure of N2C/D282C stabilized opsin bound to RS16 | | 分子名称: | 5-chloranyl-2-(2-oxidanylidene-2-spiro[1,3-benzodioxole-2,4'-piperidine]-1'-yl-ethyl)-3~{H}-pyridin-6-one, PALMITIC ACID, Rhodopsin, ... | | 著者 | Mattle, D, Standfuss, J, Dawson, R. | | 登録日 | 2018-01-23 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Ligand channel in pharmacologically stabilized rhodopsin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5J6Q
 
 | | Cwp8 from Clostridium difficile | | 分子名称: | CHLORIDE ION, Cell wall binding protein cwp8, SULFATE ION | | 著者 | Renko, M, Usenik, A, Turk, D. | | 登録日 | 2016-04-05 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
1PTT
 
 | |
1PTV
 
 | |
6Y0P
 
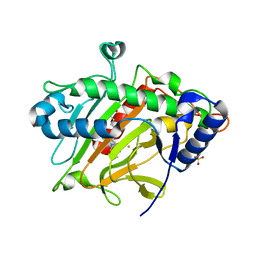 | | isopenicillin N synthase in complex with IPN and Fe using FT-SSX methods | | 分子名称: | FE (III) ION, ISOPENICILLIN N, Isopenicillin N synthase, ... | | 著者 | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | 登録日 | 2020-02-10 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
1YJ9
 
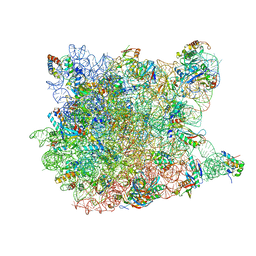 | | Crystal Structure Of The Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui Containing a three residue deletion in L22 | | 分子名称: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | 著者 | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | 登録日 | 2005-01-13 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1YJN
 
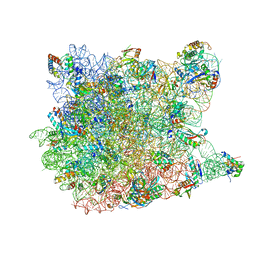 | | Crystal Structure Of Clindamycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | 分子名称: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | 著者 | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | 登録日 | 2005-01-14 | | 公開日 | 2005-04-26 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
6G5A
 
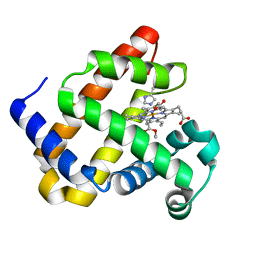 | | Heme-carbene complex in myoglobin H64V/V68A containing an N-methylhistidine as the proximal ligand, 1.48 angstrom resolution | | 分子名称: | ETHYL ACETATE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tinzl, M, Hayashi, T, Mori, T, Hilvert, D. | | 登録日 | 2018-03-29 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.483 Å) | | 主引用文献 | Capture and characterization of a reactive haem-carbenoid complex in an artificial metalloenzyme
Nat Catal, 1, 2018
|
|
1ZAN
 
 | | Crystal structure of anti-NGF AD11 Fab | | 分子名称: | CHLORIDE ION, Fab AD11 Heavy Chain, Fab AD11 Light Chain | | 著者 | Covaceuszach, S, Cattaneo, A, Cassetta, A, Lamba, D. | | 登録日 | 2005-04-06 | | 公開日 | 2006-04-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Dissecting NGF interactions with TrkA and p75 receptors by structural and functional studies of an anti-NGF neutralizing antibody.
J.Mol.Biol., 381, 2008
|
|
5HIU
 
 | | Structure of the TSC2 N-terminus | | 分子名称: | GTPase activator-like protein | | 著者 | Zech, R, Kiontke, S, Kummel, D. | | 登録日 | 2016-01-12 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of the Tuberous Sclerosis Complex 2 (TSC2) N Terminus Provides Insight into Complex Assembly and Tuberous Sclerosis Pathogenesis.
J.Biol.Chem., 291, 2016
|
|
6EPL
 
 | | Ras guanine exchange factor SOS1 (Rem-cdc25) in complex with KRAS(G12C) | | 分子名称: | GLYCEROL, GTPase KRas, Son of sevenless homolog 1 | | 著者 | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Briem, H, Petersen, K, Badock, V. | | 登録日 | 2017-10-12 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1ZJD
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Kunitz Protease Inhibitor Domain of Protease Nexin II | | 分子名称: | Catalytic Domain of Coagulation Factor XI, Kunitz Protease Inhibitory Domain of Protease Nexin II | | 著者 | Jin, L, Navaneetham, D, Pandey, P, Strickler, J.E, Babine, R.E, Walsh, P.N, Abdel-Meguid, S.S. | | 登録日 | 2005-04-28 | | 公開日 | 2005-08-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural and Mutational Analyses of the Molecular Interactions between the Catalytic Domain of Factor XIa and the Kunitz Protease Inhibitor Domain of Protease Nexin 2
J.Biol.Chem., 280, 2005
|
|
1N5D
 
 | |
