4GLU
 
 | | Crystal structure of the mirror image form of VEGF-A | | Descriptor: | ACETATE ION, D- Vascular endothelial growth factor-A, GLYCEROL, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4CDA
 
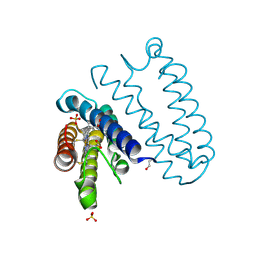 | | Spectroscopically-validated structure of ferric cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C', HEME C, SULFATE ION | | Authors: | Kekilli, D, Dworkowski, F, Antonyuk, S, Hough, M.A. | | Deposit date: | 2013-10-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fingerprinting Redox and Ligand States in Haemprotein Crystal Structures Using Resonance Raman Spectroscopy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BRX
 
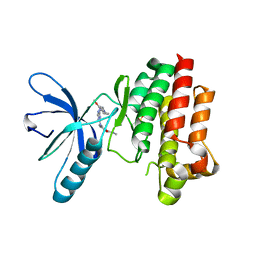 | | Focal Adhesion Kinase catalytic domain in complex with a diarylamino- 1,3,5-triazine inhibitor | | Descriptor: | 2-(4-(2-methoxy-4-morpholinophenylamino)-1,3,5-triazin-2-ylamino)-N-methylbenzamide, FOCAL ADHESION KINASE 1, SULFATE ION | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2013-06-05 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of Novel Diarylamino-1,3,5-Triazine Derivatives as Fak Inhibitors with Anti-Angiogenic Activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4BTV
 
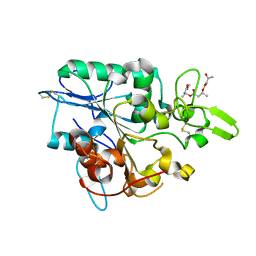 | | Structure of PhaZ7 PHB depolymerase in complex with 3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, PHB DEPOLYMERASE PHAZ7 | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-19 | | Release date: | 2013-09-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
4C28
 
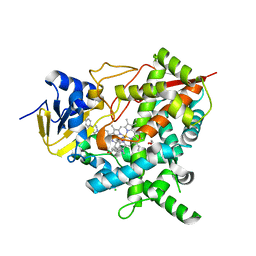 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(4-chlorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(4-chlorophenyl)piperazin-1-yl)-2-fluorobenzamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Vieira, D.F, Calvet, C.M, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2013-08-16 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Binding Mode and Potency of N-Indolyl-Oxopyridinyl-4-Amino-Propanyl-Based Inhibitors Targeting Trypanosoma Cruzi Cyp51
J.Med.Chem., 57, 2014
|
|
4BPB
 
 | | STRUCTURAL INSIGHTS INTO RNA RECOGNITION BY RIG-I | | Descriptor: | 5'-R(*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP)-3', PROBABLE ATP-DEPENDENT RNA HELICASE DDX58, SULFATE ION, ... | | Authors: | Luo, D, Pyle, A.M. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Structural Insights Into RNA Recognition by Rig-I.
Cell(Cambridge,Mass.), 147, 2011
|
|
4G2H
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | (3E,5E)-6-(3-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}phenyl)-1,1,1-trifluoro-2-(trifluoromethyl)octa-3,5-dien-2-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
4CC0
 
 | | Notch ligand, Jagged-1, contains an N-terminal C2 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN JAGGED-1, ... | | Authors: | Chilakuri, C.R, Sheppard, D, Ilagan, M.X.G, Holt, L.R, Abbott, F, Liang, S, Kopan, R, Handford, P.A, Lea, S.M. | | Deposit date: | 2013-10-17 | | Release date: | 2013-11-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Analysis Uncovers Lipid-Binding Properties of Notch Ligands
Cell Rep., 5, 2013
|
|
4EON
 
 | | Thr 160 phosphorylated CDK2 H84S, Q85M, Q131E - human cyclin A3 complex with the inhibitor RO3306 | | Descriptor: | (5E)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOR
 
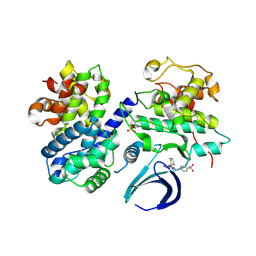 | | Thr 160 phosphorylated CDK2 WT - human cyclin A3 complex with the inhibitor NU6102 | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EQ8
 
 | | Crystal structure of PA1844 from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, Putative uncharacterized protein | | Authors: | Shang, G, Li, N, Zhang, J, Lu, D, Yu, Q, Zhao, Y, Liu, X, Xu, S, Gu, L. | | Deposit date: | 2012-04-18 | | Release date: | 2012-09-12 | | Last modified: | 2013-07-24 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Structural insight into how Pseudomonas aeruginosa peptidoglycanhydrolase Tse1 and its immunity protein Tsi1 function.
Biochem.J., 448, 2012
|
|
4E45
 
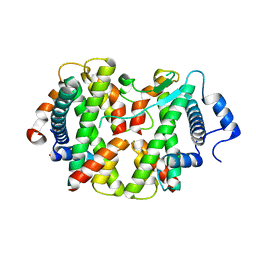 | | Crystal structure of the hMHF1/hMHF2 Histone-Fold Tetramer in Complex with Fanconi Anemia Associated Helicase hFANCM | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein, ... | | Authors: | Fox III, D, Zhao, Y, Yang, W, Weidong, W. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal that FANCM remodels the MHF Tetramer in favor of binding Branched DNA
To be Published
|
|
4GJU
 
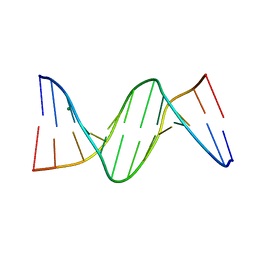 | | 5-Methylcytosine modified DNA oligomer | | Descriptor: | 5-Methylcytosine modified DNA oligomer, MAGNESIUM ION | | Authors: | Spingler, B, Renciuk, D, Vorlickova, M. | | Deposit date: | 2012-08-10 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.412 Å) | | Cite: | Crystal structures of B-DNA dodecamer containing the epigenetic modifications 5-hydroxymethylcytosine or 5-methylcytosine.
Nucleic Acids Res., 41, 2013
|
|
4EEH
 
 | | Hsp90 Alpha N-terminal Domain in Complex with an Inhibitor 3-(4-Hydroxy-phenyl)-1H-indazol-6-ol | | Descriptor: | 3-(4-hydroxyphenyl)-1H-indazol-6-ol, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Graedler, U, Buchstaller, H.-P. | | Deposit date: | 2012-03-28 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of hydroxy-indazole-carboxamides as novel small molecule inhibitors of Hsp90
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4GPK
 
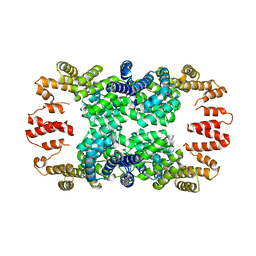 | | Crystal structure of NprR in complex with its cognate peptide NprX | | Descriptor: | NprR, NprX peptide | | Authors: | Zouhir, S, Guimaraes, B, Perchat, S, Nicaise, M, Lereclus, D, Nessler, S. | | Deposit date: | 2012-08-21 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR.
Nucleic Acids Res., 41, 2013
|
|
4EFT
 
 | | Hsp90 Alpha N-terminal Domain in Complex with an Inhibitor 3-Cyclohexyl-2-(6-hydroxy-1H-indazol-3-yl)-propionitrile | | Descriptor: | (2R)-3-cyclohexyl-2-(6-hydroxy-1H-indazol-3-yl)propanenitrile, Heat shock protein HSP 90-alpha | | Authors: | Musil, D, Lehmann, M, Graedler, U, Buchstaller, H.-P. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Fragment-based discovery of hydroxy-indazole-carboxamides as novel small molecule inhibitors of Hsp90
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EH7
 
 | | Human p38 MAP kinase in complex with NP-F6 and RL87 | | Descriptor: | (3-phenoxyphenyl)methanol, Mitogen-activated protein kinase 14, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | Authors: | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | Deposit date: | 2012-04-02 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
4H0K
 
 | | Mutant m58h of Nostoc sp cytochrome c6 | | Descriptor: | Cytochrome c6, HEME C | | Authors: | Pannu, N.S, Skubak, P, Cavazzini, D, Rossi, G.L, Ubbink, M. | | Deposit date: | 2012-09-08 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The dynamic complex of cytochrome c6 and cytochrome f studied with paramagnetic NMR spectroscopy
Biochim.Biophys.Acta, 1837, 2014
|
|
4EOL
 
 | | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with the inhibitor RO3306 | | Descriptor: | (5E)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOQ
 
 | | Thr 160 phosphorylated CDK2 WT - human cyclin A3 complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4H50
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 E175Q/T1776R/Q177G mutant (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
4BT3
 
 | | acetolactate decarboxylase with a bound (2R,3R)-2,3-Dihydroxy-2- methylbutanoic acid | | Descriptor: | (2R,3R)-2,3-Dihydroxy-2-methylbutanoic acid, ALPHA-ACETOLACTATE DECARBOXYLASE, ZINC ION | | Authors: | A Marlow, V, Rea, D, Najmudin, S, Wills, M, Fulop, V. | | Deposit date: | 2013-06-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and Mechanism of Acetolactate Decarboxylase.
Acs Chem.Biol., 8, 2013
|
|
4BZ0
 
 | | Structural characterization using Sulfur-SAD of the cytoplasmic domain of Burkholderia pseudomallei PilO2Bp, an actin-like protein component of a Type IVb R64-derivative pilus machinery. | | Descriptor: | POTASSIUM ION, PUTATIVE TYPE IV PILUS BIOSYNTHESIS PROTEIN | | Authors: | Lassaux, P, Manjasetty, B.A, Conchillo-Sole, O, Yero, D, Gourlay, L, Perletti, L, Daura, X, Belrhali, H, Bolognesi, M. | | Deposit date: | 2013-07-22 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Redefining the Pf06864 Pfam Family Based on Burkholderia Pseudomallei Pilo2BP S-Sad Crystal Structure.
Plos One, 9, 2014
|
|
4C0X
 
 | | The crystal strucuture of PpAzoR in complex with anthraquinone-2- sulfonate | | Descriptor: | 9,10-dioxo-9,10-dihydroanthracene-2-sulfonic acid, FLAVIN MONONUCLEOTIDE, FMN-DEPENDENT NADH-AZOREDUCTASE 1, ... | | Authors: | Goncalves, A.M.D, de Sanctis, D, Bento, I. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The Crystal Structure of Pseudomonas Putida Azor: The Active Site Revisited.
FEBS J., 280, 2013
|
|
4C1F
 
 | | Crystal structure of the metallo-beta-lactamase IMP-1 with L-captopril | | Descriptor: | BETA-LACTAMASE IMP-1, L-CAPTOPRIL, SULFATE ION, ... | | Authors: | Zollman, D, Brem, J, McDonough, M.A, van Berkel, S.S, Schofield, C.J. | | Deposit date: | 2013-08-12 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers.
Antimicrob. Agents Chemother., 60, 2015
|
|
