8I3X
 
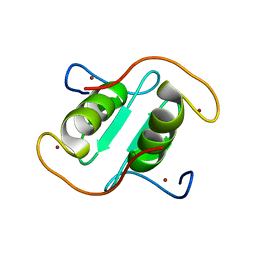 | | Rice APIP6-RING homodimer | | Descriptor: | RING-type domain-containing protein, ZINC ION | | Authors: | Zheng, Y, Zhang, X, Liu, Y, Liu, J, Wang, D. | | Deposit date: | 2023-01-18 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of rice APIP6 reveals a new dimerization mode of RING-type E3 ligases that facilities the construction of its working model
Phytopathol Res, 5, 2023
|
|
4Y0O
 
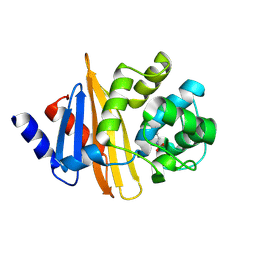 | | Crystal structure of OXA-58, a carbapenem hydrolyzing Class D beta-lactamase from Acinetobacter baumanii. | | Descriptor: | Beta-lactamase | | Authors: | Pratap, S, Katiki, M, Gill, P, Golemi-Kotra, D, Kumar, P. | | Deposit date: | 2015-02-06 | | Release date: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Active-Site Plasticity Is Essential to Carbapenem Hydrolysis by OXA-58 Class D beta-Lactamase of Acinetobacter baumannii.
Antimicrob.Agents Chemother., 60, 2015
|
|
3AQB
 
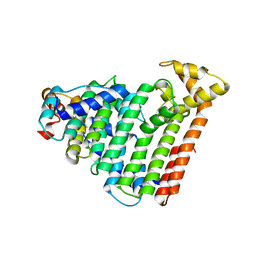 | | M. luteus B-P 26 heterodimeric hexaprenyl diphosphate synthase in complex with magnesium | | Descriptor: | CHLORIDE ION, Component A of hexaprenyl diphosphate synthase, Component B of hexaprenyl diphosphate synthase, ... | | Authors: | Sasaki, D, Fujihashi, M, Okuyama, N, Kobayashi, Y, Noike, M, Koyama, T, Miki, K. | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of heterodimeric hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 reveals that the small subunit is directly involved in the product chain length regulation.
J.Biol.Chem., 286, 2011
|
|
3DSD
 
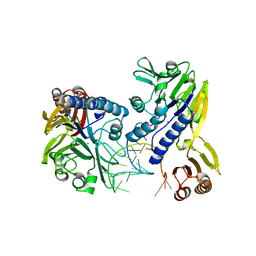 | | Crystal structure of P. furiosus Mre11-H85S bound to a branched DNA and manganese | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DAP*DCP*DAP*DAP*DGP*DCP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DGP*DTP*DGP*DGP*DAP*DTP*DA)-3'), DNA double-strand break repair protein mre11, MANGANESE (II) ION | | Authors: | Williams, R.S, Moiani, D, Tainer, J.A. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair.
Cell(Cambridge,Mass.), 135, 2008
|
|
6VM4
 
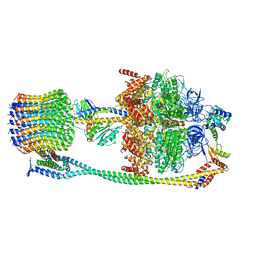 | | Chloroplast ATP synthase (C2, CF1FO) | | Descriptor: | ATP synthase delta chain, chloroplastic, ATP synthase epsilon chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.08 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
4Y2N
 
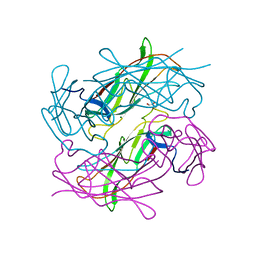 | | Structure of CFA/I pili major subunit CfaB trimer | | Descriptor: | 1,2-ETHANEDIOL, CFA/I fimbrial subunit B, MAGNESIUM ION | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2015-02-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Off-pathway assembly of fimbria subunits is prevented by chaperone CfaA of CFA/I fimbriae from enterotoxigenic E. coli.
Mol. Microbiol., 102, 2016
|
|
4XNV
 
 | | The human P2Y1 receptor in complex with BPTU | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2-(2-tert-butylphenoxy)pyridin-3-yl]-3-[4-(trifluoromethoxy)phenyl]urea, CHOLESTEROL, ... | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
7QEP
 
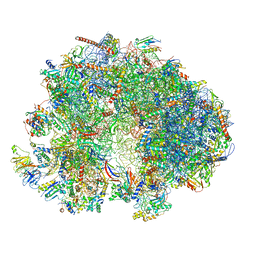 | | Cryo-EM structure of the ribosome from Encephalitozoon cuniculi | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Nicholson, D, Ranson, N.A, Melnikov, S.V. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Adaptation to genome decay in the structure of the smallest eukaryotic ribosome
Nat Commun, 13, 2022
|
|
3AVX
 
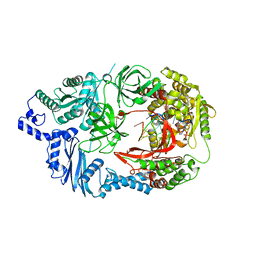 | | Structure of viral RNA polymerase complex 5 | | Descriptor: | 3'-DEOXY-GUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AQN
 
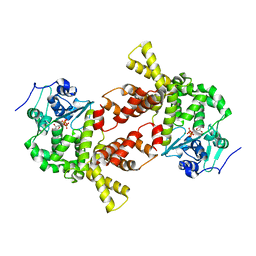 | |
4Y2L
 
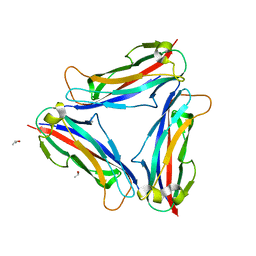 | | Structure of CFA/I pili major subunit CfaB trimer | | Descriptor: | CALCIUM ION, CFA/I fimbrial subunit B, ISOPROPYL ALCOHOL | | Authors: | Bao, R, Xia, D. | | Deposit date: | 2015-02-10 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.746 Å) | | Cite: | Off-pathway assembly of fimbria subunits is prevented by chaperone CfaA of CFA/I fimbriae from enterotoxigenic E. coli.
Mol. Microbiol., 102, 2016
|
|
6PQ0
 
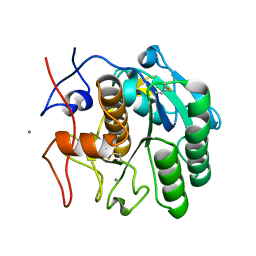 | | LCP-embedded Proteinase K treated with MPD | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
3AQL
 
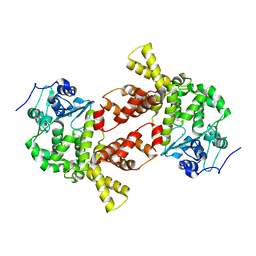 | |
6PPF
 
 | | Bacterial 45SRbgA ribosomal particle class B | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Ortega, J, Seffouh, A, Jain, N, Jahagirdar, D, Basu, K, Razi, A, Ni, X, Guarne, A, Britton, R.A. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
6PRK
 
 | | X-ray Crystal Structure of Bacillus subtilis RicA in complex with RicF | | Descriptor: | RicA, RicF | | Authors: | Khaja, F.T, Jeffrey, P.D, Neiditch, M.B, Dubnau, D. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Function Studies of the Bacillus subtilis Ric Proteins Identify the Fe-S Cluster-Ligating Residues and Their Roles in Development and RNA Processing.
Mbio, 10, 2019
|
|
6PQ4
 
 | | LCP-embedded Proteinase K treated with lipase | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Bu, G, Zhu, L, Jing, L, Shi, D, Gonen, T, Liu, W, Nannenga, B.L. | | Deposit date: | 2019-07-08 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | Structure Determination from Lipidic Cubic Phase Embedded Microcrystals by MicroED.
Structure, 28, 2020
|
|
3ATQ
 
 | | Geranylgeranyl Reductase (GGR) from Sulfolobus acidocaldarius | | Descriptor: | Conserved Archaeal protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, TETRADECANE | | Authors: | Sasaki, D, Fujihashi, M, Murakami, M, Yoshimura, T, Hemmi, H, Miki, K. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and mutation analysis of archaeal geranylgeranyl reductase
J.Mol.Biol., 409, 2011
|
|
6PRH
 
 | |
3AX3
 
 | |
3AX2
 
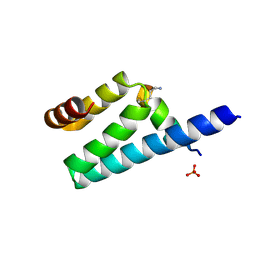 | | Crystal structure of rat TOM20-ALDH presequence complex: a disulfide-tethered complex with a non-optimized, long linker | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-28 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
4Y25
 
 | | Bacterial polysaccharide outer membrane secretin | | Descriptor: | Poly-beta-1,6-N-acetyl-D-glucosamine export protein | | Authors: | Wang, Y, AndolePannuri, A, Ni, D, Zhou, H, Cao, X, Lu, X, Romeo, T, Huang, Y. | | Deposit date: | 2015-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Structural Basis for Translocation of a Biofilm-supporting Exopolysaccharide across the Bacterial Outer Membrane
J.Biol.Chem., 291, 2016
|
|
3AX5
 
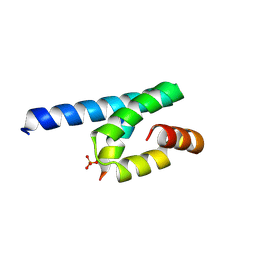 | | Crystal structure of rat TOM20-ALDH presequence complex: A complex (form1) between Tom20 and a disulfide-bridged presequence peptide containing D-Cys and L-Cys at the i and i+3 positions. | | Descriptor: | Aldehyde dehydrogenase, mitochondrial, Mitochondrial import receptor subunit TOM20 homolog, ... | | Authors: | Saitoh, T, Maita, Y, Kohda, D. | | Deposit date: | 2011-03-29 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic snapshots of tom20-mitochondrial presequence interactions with disulfide-stabilized peptides.
Biochemistry, 50, 2011
|
|
8I5K
 
 | |
8I5J
 
 | |
8HZC
 
 | |
